Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer
Affiliations.
- 1 Laboratory of Virology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, MT 59840, USA; Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Oxford OX3 7BN, UK.
- 2 Laboratory of Virology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, MT 59840, USA; Marshall University Joan C. Edwards School of Medicine, Huntington, WV 25701, USA.
- 3 Laboratory of Virology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, MT 59840, USA.
- 4 Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Oxford OX3 7BN, UK.
- 5 Research Technologies Branch, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, MT 59840, USA.
- 6 Department of Medicine, University of Washington, Seattle, WA 98195, USA.
- 7 EvergreenHealth, Kirkland, WA 98034, USA. Electronic address: [email protected].
- 8 Laboratory of Virology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, MT 59840, USA. Electronic address: [email protected].
- PMID: 33248470
- PMCID: PMC7640888
- DOI: 10.1016/j.cell.2020.10.049
Long-term severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) shedding was observed from the upper respiratory tract of a female immunocompromised individual with chronic lymphocytic leukemia and acquired hypogammaglobulinemia. Shedding of infectious SARS-CoV-2 was observed up to 70 days, and of genomic and subgenomic RNA up to 105 days, after initial diagnosis. The infection was not cleared after the first treatment with convalescent plasma, suggesting a limited effect on SARS-CoV-2 in the upper respiratory tract of this individual. Several weeks after a second convalescent plasma transfusion, SARS-CoV-2 RNA was no longer detected. We observed marked within-host genomic evolution of SARS-CoV-2 with continuous turnover of dominant viral variants. However, replication kinetics in Vero E6 cells and primary human alveolar epithelial tissues were not affected. Our data indicate that certain immunocompromised individuals may shed infectious virus longer than previously recognized. Detection of subgenomic RNA is recommended in persistently SARS-CoV-2-positive individuals as a proxy for shedding of infectious virus.
Keywords: COVID-19; SARS-CoV-2; asymptometic; chronic lymphocytic leukemia; convalescent plasma; immunocompromised; infectious virus; long-term shedding; within host evolution.
Published by Elsevier Inc.

Publication types
- Case Reports
- Research Support, N.I.H., Intramural
- Research Support, Non-U.S. Gov't
- Antibodies, Viral / blood
- Antibodies, Viral / immunology
- COVID-19 / complications
- COVID-19 / immunology*
- COVID-19 / virology
- Common Variable Immunodeficiency / blood
- Common Variable Immunodeficiency / complications
- Common Variable Immunodeficiency / immunology*
- Common Variable Immunodeficiency / virology
- Leukemia, Lymphocytic, Chronic, B-Cell / blood
- Leukemia, Lymphocytic, Chronic, B-Cell / complications
- Leukemia, Lymphocytic, Chronic, B-Cell / immunology*
- Leukemia, Lymphocytic, Chronic, B-Cell / virology
- Respiratory Tract Infections / blood
- Respiratory Tract Infections / complications
- Respiratory Tract Infections / immunology
- Respiratory Tract Infections / virology
- SARS-CoV-2 / immunology
- SARS-CoV-2 / isolation & purification*
- SARS-CoV-2 / pathogenicity
- Antibodies, Viral
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- MR/S007555/1/MRC_/Medical Research Council/United Kingdom
- T32 GM008169/GM/NIGMS NIH HHS/United States
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Wiley - PMC COVID-19 Collection

COVID‐19: Virology, biology and novel laboratory diagnosis
Malihe mohamadian.
1 Department of Molecular Medicine, Birjand University of Medical Sciences, Birjand Iran
2 Cellular and Molecular Research Center, Birjand University of Medical Sciences, Birjand Iran
Hossein Chiti
3 Zanjan Metabolic Diseases Research Center, Zanjan University of Medical Sciences, Zanjan Iran
Alireza Shoghli
4 Social Determinants of Health Research Center, Zanjan University of Medical Sciences, Zanjan Iran
Sajjad Biglari
5 School of Medicine, Zanjan University of Medical Sciences, Zanjan Iran
Negin Parsamanesh
Abdolreza esmaeilzadeh.
6 Department of Immunology, Zanjan University of Medical Science, Zanjan Iran
7 Cancer Gene Therapy Research Center, Zanjan University of Medical Science, Zanjan Iran
Associated Data
Data sharing is not applicable to this review because no datasets were generated or analyzed during the present study.
At the end of December 2019, a novel coronavirus tentatively named SARS‐CoV‐2 in Wuhan, a central city in China, was announced by the World Health Organization. SARS‐CoV‐2 is an RNA virus that has become a major public health concern after the outbreak of the Middle East Respiratory Syndrome‐CoV (MERS‐CoV) and Severe Acute Respiratory Syndrome‐CoV (SARS‐CoV) in 2002 and 2012, respectively. As of 29 October 2020, the total number of COVID‐19 cases had reached over 44 million worldwide, with more than 1.17 million confirmed deaths.
SARS‐CoV‐2 infected patients usually present with severe viral pneumonia. Similar to SARS‐CoV, the virus enters respiratory tract cells via the angiotensin‐converting enzyme receptor 2. The structural proteins play an essential role in budding the virus particles released from different host cells. To date, an approved vaccine or treatment option of a preventive character to avoid severe courses of COVID‐19 is still not available.
Conclusions
In the present study, we provide a brief review of the general biological features of CoVs and explain the pathogenesis, clinical symptoms and diagnostic approaches regarding monitoring future infectivity and prevent emerging COVID‐19 infections.
The essential structural proteins play an important role in budding the virus particles released from different host cells. Notably, reported cases confirm human‐to‐human transmission, along with numerous cases of exported virus infections all over the world. However, to date, an approved vaccine or treatment option of preventive character to avoid severe courses of COVID‐19 is still not available. In the present study, we provide a brief review of the general biological features of CoVs and explain the pathogenesis, clinical symptoms and diagnostic approaches with regard to monitoring future infectivity and preventing emerging COVID‐19 infections.

1. INTRODUCTION
In December 2019, a novel coronavirus (nCoV) termed “SARS‐CoV‐2”, announced by the World Health Organization (WHO) as being responsible for the outbreak of COVID‐19, was
reported. 1 , 2 The incidence of the SARS‐CoV (Severe Acute Respiratory Syndrome‐coronavirus) in 2002 and 2003 and the MERS‐CoV (Middle East Respiratory Syndrome‐coronavirus) in 2012 showed the potential for the transmission of newly emerging CoVs from animal to human and person to person. 3 , 4 , 5 In total, seven human coronaviruses (HCoVs) have now been discovered, including HCoV229E, HCoV‐OC43, HCoV‐NL63, HKU1, SARS‐CoV, MERS‐CoV and SARS‐CoV‐2. 6 , 7
In the last two decades, SARS‐CoV and MERS‐CoV have caused epidemics with mortality rates of approximately 9.5% and 34.4%, respectively. 8 COVID ‐ 19 was the third highly epidemic disease to be detected, with a lower mortality rate than SARS and MERS, differing from country to country. According to WHO statistics, there are 45,678,440 (1 November 2020) confirmed cases in 219 countries caused by the high transmission capacity of SARS‐CoV‐2. 9 Hence, to characterize the acute infection in humans as a result of the SARS‐CoV‐2, scientists and governments have urgently taken decisive steps to monitor its outbreak and carry out etiological research. 10 In combination with advances in molecular technologies, the development of a viable vaccine appears to be imminent. 11 , 12
The higher transmissibility, varied clinical manifestations and lower pathogenicity of COVID‐19 could be a result of diversity in the biology and genome structure of the SARS‐CoV‐2 compared to SARS‐CoV and MERS‐CoV. 13 , 14 This review focuses on the virus biology, clinical symptoms and potential diagnostic routes for achieve efficient prevention and reduction of COVID‐19 mortality.
2. GENOMICS STRUCTURE AND BIOLOGICAL FEATURES OF SARS‐COV‐2
Coronaviruses belong to the order Nidovirales in the family coronaviridae. Coronavirinae and Torovirinae subfamilies are divided from the family. The subfamily Coronavirinae is further divided into four genera: Alpha‐ , Beta‐ , Gamma‐ and Deltacoronavirus . 15 Phylogenic analysis revealed that SARS‐CoV‐2 is closely related to the beta‐coronaviruses. Similar to other coronaviruses, the genome of SARS‐CoV‐2 is positive‐sense single‐stranded RNA [(+) ssRNA] with a 5′‐cap, 3'‐UTR poly(A) tail. The length of the SARS‐CoV‐2 genome is less than 30 kb, in which there are 14 open reading frames (ORFs), encoding non‐structural proteins (NSPs) for virus replication and assembly processes, structural proteins including spike (S), envelope (E), membrane/matrix (M) and nucleocapsid (N), and accessory proteins. 16 , 17 The first ORF contains approximately 65% of the viral genome and translates into either polyprotein pp1a (nsp1–11) or pp1ab (nsp1–16). Among them, six nsps (NSP3, NSP9, NSP10, NSP12, NSP15 and NSP16) play critical roles in viral replication. Other ORFs encode structural and accessory proteins. 18 , 19 The S protein is a transmembrane protein that facilitates the binding of viral envelop to angiotensin‐converting enzyme 2 (ACE2) receptors expressed on host cell surfaces. Functionally, the spike protein is composed of receptor binding (S1) and cell membrane fusion (S2) subunits. 20 The N protein attaches to the viral genome and is involved in RNA replication, virion formation and immune evasion. The nucleocapsid protein also interacts with the nsp3 and M proteins. 21 The M protein is one of the most abundant and well‐conserved proteins in the virion structure. This protein promotes the assembly and budding of viral particles through interaction with N and accessory proteins 3a and 7a. 22 , 23 The E protein is the smallest component in the SARS‐CoV‐2 structure that facilitates the production, maturation and release of virions. 18
The most complex component of the coronaviruses genome is the receptor‐binding domain (RBD) in the spike protein. 24 , 25 Six RBD amino acids are necessary for attaching to the ACE2 receptor and hosting SARS‐CoV‐like coronaviruses. According to multiple sequence alignment, they are Y442, L472, N479, D480, T487 and Y4911 in SARS‐CoV, and L455, F486, Q493, S494, N501 and Y505 in SARS‐CoV‐2. 26 Therefore, SARS‐CoV‐2 and SARS‐CoV vary with respect to five of these six residues. The SARS‐CoV strain genome sequences derived from humans were very close to those in bats. Even so, several differences have been identified between the gene sequences of the S gene and the ORF3 and ORF8 gene sequences that encode the attachment and fusion proteins and replication proteins, respectively. 27 Specific MERS‐CoV strains derived from camels were shown to be identical to those extracted from humans, with the exception of differences between the genomic regions S, ORF4b and ORF3. 28 In addition, genome sequencing‐based experiments have shown that human MERS‐CoV strains are phylogenetically linked to those of bats. However, for the S proteins, the species have a similar genome and protein structures. 29 Based on the recombination studies of orf1ab and S encoding genes, the MERS‐CoV was derived from the interchange of genetic elements between coronaviruses in camels and bats. In comparison, with a 96% overall identification, the primary protease is strongly protected among SARS‐CoV‐2 and SARS‐CoV. 29 , 30 , 31
The ACE2 protein is found in many mammalian body tissues, primarily in the lungs, kidneys, gastrointestinal tract, heart, liver and blood vessels. 32 ACE2 receptors are vital elements in regulating the renin‐angiotensin‐aldosterone system pathway. Based on structural experiments and biochemical studies, SARS‐CoV‐2 appears to have an RBD that strongly binds to ACE2 receptors of humans, cats, ferrets and other organisms with the homologous receptors. 33
The genome sequencing of SARS‐CoV‐2 in January 2020 was shown to be 96% identical to the bat coronavirus (BatCoV) RaTG13 genome and 80% identical to the SARS‐CoV genome. 34 However, significant differences exist. For example, the protein 8a sequence in the SARS‐CoV genome is absent in the 2019‐nCoV, and the protein 8b sequence of SARS‐CoV‐2 is 37 amino acids longer than that in SARS‐CoV. 35
Alignment sequence analysis of the CoV genome indicates non‐structural and structural proteins being 60% and 45% identical, respectively, among various types of CoVs. 36 These data show that nsps are more conservative than structural proteins. RNA viruses have a higher mutational load as a result of shorter replication times (Figure 1 ) . 36 Based on comparative genomic studies between SARS‐CoV‐2 and SARS‐like coronaviruses, there are 380 amino acid substitutions in the nsps genes and 27 mutations in genes encoding the spike protein S of SARS‐CoV‐2. These variations might explain the different behavioral patterns of SARS‐CoV‐2 compared to SARS‐CoVs. 8 For example, the primary N501 T mutation in the Spike protein of SARS‐CoV‐2 could have significantly increased its binding affinity to ACE2. 37

The schematic genomic structure of coronavirus. (a) COVID‐19 . (b) MERS‐CoV. (c) SARS‐CoV. The typical coronavirus genome is a single‐stranded, which is approximately 25–32 kb. It contains 5' caps and 3'‐UTR tails. (d) Coronavirusencoding structural proteins four structural genes, including spike, envelope, membrane and nucleocapsid genes, as well as accessory proteins ( 3a , 3b , 6 , 7a , 7b , 8b , 9b and ORFs)
2.1. Pathogenesis of SARS‐CoV‐2
The entry of the SARS‐CoV‐2 into host cells and release their genomes into target cells is dependent on a sequence of steps. The virus uses the protein spike, which is important for assessing tropism and virus transmissibility. Additionally, SARS‐CoV‐2 even targets human respiratory epithelial cells with ACE2 receptors, indicating a structure of RBD similar to SARS‐CoV. 38 After attachment of the S1‐RBD to the ACE2 receptor, host cell‐surface proteases such as TMPRSS2 (transmembrane serine protease 2) act on a critical cleavage site on S2. 38 This results in membrane fusion and viral infection. Following virus entry, the uncoated genomic RNA is translated into polyproteins (pp1a and pp1ab) and then assembled into replication/transcription complexes with virus‐induced double‐membrane vesicles (DMVs). Subsequently, this complex replicates and synthesizes a nested set of subgenomic RNA by genome transcription, encoding structural proteins and some accessory proteins. Newly formed virus particles are assembled by mediating the endoplasmic reticulum and the Golgi complex. Finally, virus particles are budded and released into the extracellular milieu compartment. Thus, both the viral replication cycle and progression begin. 10
Inside the host cells, survival of SARS CoVs is maintained by multiple strategies to evade the host immune mechanism, which can also be generalized to SARS‐CoV‐2. 39 , 40 As a result of the lack of pathogen‐associated molecular patterns on DMVs originating from the first step of SARS‐CoVs infection, they are not recognized by pattern recognition receptors of the host immune system. 25 Nsp1 can impede the interferon (IFN)‐I responses through several mechanisms, such as a silencing of the host translational system, the induction of host mRNA degradation and the repression of transcription factor signal transducer and activator of transcription (STAT)1 phosphorylation. Nsp3 antagonizes interferon and cytokine production by blocking the phosphorylation of interferon regulation factor 3 (IRF3) and interrupting the nuclear factor‐kappa B (NF‐ΚB) signaling pathway. NSPs 14 and 16 cooperate to form a viral 5′ cap similar to that of the host. Thus, the viral RNA genome is not recognized by immune system cells. 41 The accessory proteins ORF3b and ORF6 can disrupt the IFN signaling pathway by inhibiting IRF3 and NF‐KB‐dependent IFNβ expression and blocking the JAK‐STAT signaling pathway, respectively. Also, IFN signaling is flattened by structural proteins M and N, which result in a disturbance in TANK‐binding kinase 1 (TBK1)/IKB kinase ε and TRAF3/6‐TBK1‐IRF3/NF‐ΚB/AP1 signals. 22 , 39 Because the D614 G mutation is found in the outer spike protein of the virus, this attracts a huge amount of attention from the human immune system and may therefore impair the ability of SARS‐CoV‐2 to avoid vaccine‐induced immunity. D614 G is not in the RBD, although it is involved in the interaction between individual spike protomers that regulate their mature trimeric form on the surface of the virion by hydrogen bonding. 42 Korber et al . reported that the SARS‐CoV‐2 variant in the D614 G spike protein has become influential across the globe. Although clinical and in vitro evidence indicate that D614 G alters the phenotype of the virus, the effect of the mutation on replication, pathogenesis, vaccine and therapy development is relatively unknown. 43 From in vitro and clinical evidence, it is apparent that D614 G has a distinct phenotype, although it is not clear whether this is the outcome of verified adaptation to human ACE2, as well as whether it enhances transmissibility, or will have a significant impact. 43
2.2. Diagnosis of COVID‐19
Early diagnosis and isolation of suspected patients play a vital role in controlling this outbreak. 44 The specificity and sensitivity of different diagnostic techniques differs between populations and the types of equipment employed. 45 Several proceedures have been recommended for the diagnosis of COVID‐19:
- Clinical presentation
COVID‐19 symptoms are observed approximately 5 days after incubation. 46 The median time of symptom onset from COVID‐19 incubation is 5.1 days, and those infected display symptoms for 11.5 days. 47 This duration was shown to have a close link with the patient's immune system and age. Gastrointestinal symptoms include diarrhea, vomiting and anorexia, recorded in almost 40% of patients. 48 , 49 Up to 10% of patients with gastrointestinal symptoms show no signs of fever or respiratory tract infections. 50 COVID‐19 has also been linked to hypercoagulable disease, elevating the risk of venous thrombosis. 51 There are also records of neurological symptoms (such as fatigue, dizziness and disturbed awareness), ischemic and hemorrhagic strokes, and muscle damage. 52 Many extrapulmonary symptoms comprise skin and eye manifestations. Italian researchers have identified skin manifestations in 20% of patients. 53 The clinical outlook for children can progressively worsen as a result of respiratory failure, which could not be corrected within 1–3 days by traditional oxygen (i.e. nasal catheter 54 ) in severe cases; the hallmarks are septic shock, sepsis, extreme and continuum bleeding as a result of coagulation abnormalities, and metabolic acidosis. 55
Septic shock could cause severe damage and impair several organs, in addition to a severe pulmonary infection. When extrapulmonary system dysfunction occurs, including the circulatory and the digestive systems, septic shock is probable, and the mortality rate increases substantially. 55 Premature delivery and intrauterine hypoxia occur when the prenate is deprived of an adequate environment of oxygen. Insidious symptoms require specific care in some newborn and preterm infants. Reports have indicated a good prognosis for children within 1 or 2 weeks. 55 Children are prone to a hyperinflammatory response to COVID‐19 similar to Kawasaki disease, which responds well to management, for which a new term is being coined. 56
Also, considerable research has revealed the age distribution of adolescent patients between the ages of 25 to 89 years. Many elderly patients were between 35 and 55 years, and fewer cases among newborns and infants were found. An analysis of the initial transmission dynamics of the virus showed that the median age of patients was 59 years, varying from 15 to 89 years; most (59%) were male. 48
- Nonspecific screening tests for COVID‐19 in exposed patients
The findings of most blood tests are usually nonspecific but could help determine the causes of the disease. A complete blood count typically shows a normal or low count of white blood cells and lymphopenia. C‐reactive protein (CRP) and erythrocyte sedimentation rate were generally increased, which would optimally be rechecked on days 3, 5 and 7 after admission. 1 , 57 , 58 Creatine kinase plus myoglobin, aspartate aminotransferase and alanine aminotransferase, lactate dehydrogenase, D‐dimer, and creatine phosphokinase levels could be increased in severe forms of COVID‐19 disease. During viral‐bacterial co‐infections, procalcitonin levels may be elevated. 59 , 60 In a systematic review and meta‐analysis study, Pormohammad et al . 61 investigated the accessible laboratory results obtained among 2361 SARS‐CoV2 patients, with the results demonstrating 26% leukopenia, 13.3% leukocytosis and 62.5% lymphopenia. Also, among 2200 patients, 91% and 81% revealed elevated platelets (thrombocytosis) and CRP, respectively. 61 Additionally, a review of case studies identified clinical diagnosis and clinical parameter modification in a 47‐year‐old man diagnosed with the disease from Wuweian. 62
To investigate the effect of the coronavirus during the acute phase of the disease, plasma cytokines/chemokines tumor necrosis factor (TNF)‐α and interleukin (IL)‐1β , IL1RA, IL2, IL4, IL5, IL‐6, IL‐10, IL13, IL15 and IL17A were measured. 1 , 63 One study showed that macrophages and dendritic cells play crucial roles in an adaptive immune system. These cells contain inflammatory cytokines and chemokines, such as IL‐6, IL‐8, IL‐12, TNF‐α, monocyte chemoattractant protein‐1, granulocyte‐macrophage colony‐stimulating factor and granulocyte colony‐stimulating factor. These inflammatory reactions could cause a systemic inflammation. 64
Therefore, fecal and urine tests have been recommended for patients and health staff to detect possible alternate transmission. Consequently, the advancement of tools for determining the different transmission modes, including fecal and urine samples, is urgently warranted to develop strategies for inhibiting and minimizing transmission, as well as develop therapies to control the disease. 65
- Radiological findings
Chest X‐ray examination may display diverse imaging characteristics or patterns in COVID‐19 patients with a different disease severity and duration. Imaging results differ based on patient age, disease stage during screening, immune competency and drug therapy protocols. 66 On the other hand, computed tomagraphy (CT) imaging is essential for monitoring disease progression and assessing therapeutic effectiveness. It can be re‐examined 1 to 2 days after admission, based on the Diagnostic and Treatment Protocols Regulation (DTPR). 67
The cardinal hallmark of COVID‐19 was multiple, bilateral, posterior and peripheral ground‐glass opacities with or without pulmonary consolidation and, in severe cases, infiltrating shadows. 68 Autopsy analysis of a COVID‐19 patient displayed fluid accumulation and hyaline membrane formation in alveolar walls, which may be the primary pathological driver of the ground‐glass opacity. 69
However, further studies indicated that small patchy shadows, pleural changes, a subpleural curvilinear line and reversed halo signs are generally observed in COVID‐19 patients. 70 , 71 The intralobular lines and thickened interlobular septa were shown in a crazy‐paving pattern on the ground‐glass opacity background. 67 Also, several lobar lesions can be found in the respiratory system in children with a severe infection. Evidence showed that chest CT manifestations (pulmonary edema) reported for COVID‐19 are generally close to SARS and MERS. 69
Evidence has indicated that an initial chest CT has a higher detection rate (approximately 98%) compared to reverse transcriptase‐polymerase chain reaction (RT‐PCR) (approximately 70%) in infected patients. Xie et al . 72 demonstrated that about 3% of patients have no primary positive RT‐PCR but have a positive chest CT; therefore, both tests are recommended for COVID‐19 patients. CT of the chest comprises an urgent and simple method for detecting initial COVID‐19 infection with a high sensitivity for prompt diagnosis and disease progression monitoring in patients. Particular notice should be paid to the role of radiologists in finding novel infectious diseases.
- Molecular diagnosis
The clinical diagnosis of COVID‐19 is focused primarily on epidemiological data, clinical symptoms and some adjuvant technologies, such as nucleic acid detection and immunological assays. In addition, the isolation of SARS‐CoV‐2 requires high‐throughput equipment (biosafety level‐3) to ensure personnel safety. Moreover, serological tests have not yet been validated. In the field of molecular diagnosis, there are three main issues: (i) decreasing the number of false negatives by detecting minimal amounts of viral RNA; (ii) avoiding the number of false positives through the correct differentiation of positive signals between different pathogens; and (iii) a high capacity for fast and accurate testing of a large number of samples in a short time. 73
2.3. Nucleic acid detection
Two widely used technologies for SARS‐CoV‐2 nucleic acid detection are the real‐time RT‐PCR (rRT‐PCR) and high‐throughput sequencing. Nevertheless , as a result of a reliance on equipment and high costs, high‐throughput sequencing in clinical diagnosis is restricted. Access to the whole genome structure of SARS‐CoV‐2 has helped the design of specific primers and has introduced the best diagnostic protocols. 47 , 74 In the first published reports on applying the rRT‐PCR in COVID‐19 diagnosis, targeting the spike gene region (S) of SARS‐COV‐2 has shown remarkable specificity and limited sensitivity. 68 Later, the sensitivity of this technique was greatly improved by the use of specific probes for the other viral‐specific genes, including RNA‐dependent RNA polymerase ( RdRp ) in the ORF1ab region, Nucleocapsid ( N ) and Envelop ( E ). To avoid cross‐reaction with other human coronaviruses and prevent the potential genetic drift of SARS‐CoV‐2, two molecular targets should be involved in this assay: one nonspecific target to detect other CoVs, and one specific target for SARS‐CoV‐2. The comparison of the results obtained from targeting all studied genes exhibited that the RdRp gene is the most appropriate target with the highest sensitivity. The RdRp assays were validated in approximately 30 European laboratories using synthetic nucleic acid technology. 73 Currently, Chan et al . 75 have proposed a novel RT‐PCR assay targeting a sequence of the RdRp/Hel that could detect low SARS‐CoV‐2 load in the upper respiratory tract, plasma and saliva samples without any cross‐reactivity with other common respiratory viruses. Although the CDC‐recommended assays in the USA rely on two nucleocapsid proteins N1 and N2, the WHO recommends the E gene assay as a first‐line screening, followed by the RdRp gene assay as a confirmatory test. Based on the most recent evidence, the QIAstat‐Dx SARS‐CoV‐2 panel, a multiplex RT‐real time PCR system targeting genes RdRp and E, remains highly sensitive despite the nucleotide variations affecting the annealing of the PCR assay. 76
Generally, quantitative (RT‐PCR) RT‐qPCR has high specificity as a gold standard assay for the final diagnosis of COVID‐19. However, its sensitivity could be variable based on viral load, RNA extraction technique, sampling source and disease stage during the time of sampling. Indeed, the RT‐PCR false‐positive results are related to the cross‐contamination of samples and handling errors. By contrast, inaccuracies during any stage of the collection, storage and processing of samples may lead to false‐negative results. Some studies have revealed that samples from the upper respiratory tract (bottom of the nostrils and the oropharynx) are more desirable for the RT‐PCR assay as a result of many viral copies. 77 Moreover, other shortcomings of RT‐qPCR assays include biological safety hazards arising from maintaining and working on patient samples, as well as time‐consuming and cumbersome nucleic acid detection process. 66 , 68
To improve the molecular diagnostic techniques for COVID‐19, isothermal amplification‐based methods are currently in development. The loop‐mediated isothermal amplification (LAMP) utilizes the DNA polymerase and 4 to 6 different primers binding to the distinct sequences on the target genome. 78 In the LAMP reactions, the amplified DNA is indicated by turbidity arising from a by‐product of the reaction, a detectable color generated by a pH‐sensitive dye, or fluorescence produced by a fluorescent dye. 79 The approach occurs at a single temperature, in less than 1 hour, and with minimal background signals. The LAMP diagnostic testing for COVID‐19 is more specific and sensitive compared to the conventional RT‐PCR assays and does not dependent on specialized laboratory equipment such as a thermocycler. However, as a result of the multiplicity of primers used in this method, optimizing the reaction conditions presents a major challenge. 80 , 81
2.4. Microarray‐based technique
Microarray is a rapid and high‐throughput method for the COVID‐19 assay. 82 As a brief summary of the protocol, the coronavirus RNA will first produce cDNA labeled with specific probes via reverse transcription. Complementary DNA is produced by coronavirus RNA templates and then through reverse transcription labeling with particular probes. The labeled targets are hybridized to the probe microarray. Free DNAs are removed by washing the solution. Finally, particular probes identify COVID‐19 RNA. 82 Shi et al . 83 successfully performed SARS‐CoV detection in samples from patients. In their study, Xu et al . 84 investigated a wide range of spike gene polymorphisms with great accuracy. Also, other studies have designed fluorescence and nonfluorescence methods to detect the entire coronavirus genus with promising efficacy. 85 , 86 Jiang et al . 87 constructed a SARS‐CoV‐2 proteome microarray consisting of 18 out of 28 expected proteins and administered it to 29 convalescent cases to characterize the immunoglobulin (Ig)G and IgM reactions in the sera. It was revealed that all of these patients had IgM and IgG antibodies, which recognize and bind SARS‐CoV‐2 proteins, especially S1and N proteins. In addition to these proteins, important antibody responses to NSP5 and ORF9b are also recognized. The S1‐specific IgG signal relates strongly to age and lactate dehydrogenase lactate dehydrogenase levels and negatively relates to the lymphocyte ratio. Shen et al . progressed the RT‐LAMP experiment to show signals using a quenching probe with the same efficiency as the standard RT‐PCR test with respect to MERS‐CoV identification. 80
- Immunological diagnosis
Antigen detection and immunological techniques can be used for a rapid and cost‐effective diagnosis at the same time as providing an alternative to molecular methods. Immunological techniques including the immunofluorescence assay, direct fluorescence antibody test, nucleocapsid protein detection assay, protein chip, semiconductor quantum dots and the microneutralization assay define a binding between a viral antigen and a specific antibody. 88 , 89 , 90 , 91 These immunological methods are simple to operate but have low specificity/sensitivity. In the case of COVID‐19, virus morphology can be observed by electron microscopy according to traditional Koch’s postulates. 92 , 93 Serological tests can improve coronavirus detection such that associated antigens and monoclonal antibodies can represent a new diagnostic approach for future development (Figure 2 ). 94 , 95

Diagnostic protocol recommended for COVID‐19
Serological tests could be specific to one type of immunoglobulin, they can concurrently measure IgM and IgG antibodies, or they may be absolute antibody examinations, which often measure IgA antibodies. 96 Based on the specific procedure and device, these experiments will usually be carried out within 1–2 hours after a sample arrives in the laboratory and is loaded onto the appropriate platform. 97 Guo et al . 98 indicated that IgA and IgM antibodies have positive rates of 93.0% and 85.5% after 3–6 days, respectively. Also, 78.0% of positive IgG antibodies were detected during 10–18 days. The efficiency of detection by an IgM enzyme‐linked immunposorbent assay (ELISA) is higher than that of qPCR after 5.5 days of symptom onset. After 5 days, IgM ELISA detection is more efficient than a qPCR.
Moreover, the combination of PCR and IgM ELISA increased the detection rate by 98.5%. 98 Xiang et al . 99 tested 63 infected patients of SARS‐CoV‐2 admitted to Jinyintan Hospital in Wuhan, Hubei, China. Patient serum samples were evaluated using an ELISA and indirect ELISA IgG capture. The study results indicate that IgM was positive with an accuracy of 64.3%, a sensitivity of 44.4% and a specificity of 100% in 28 of 63 samples. The sample identification of 52 cases also showed a positive IgG test with a sensitivity of 82.54%, a specificity of 100% and an accuracy of 88.8%. In addition, a sensitivity of 87.3% was achieved using IgM and IgG combination analysis. 99
Liu et al . evaluated the anti‐IgM and anti‐IgG produced against recombinant spike protein and nucleocapsid protein of SARS‐CoV‐2 in 397 PCR confirmed COVID‐19 patients and 128 negative cases at eight distinct clinical sites. The average sensitivity and specificity of the examination was 88.5% and 90.5%, respectively. The findings showed considerable detection consistency among the different types of venous and fingerstick blood samples. Compared to a single IgM or IgG test, the IgM‐IgG combination analysis has a higher effectiveness and sensitivity. 37 , 100 Therefore, it is important and urgent to improve several sensitive and specific supplementary approaches for COVID‐19 diagnosis.
- Novel techniques
2.5. CRISPR technique
Nucleic acid detection with CRISPR‐Cas13a/C2c2 is a highly rapid, sensitive and specific molecular detection platform, which may aid in the epidemiology, diagnosis and control of the disease. In addition, Cas13a/C2c2 can detect the expression of transcripts in live cells and different diseases. 101 , 102 Zhang et al. presented a protocol for the detection of COVID‐19 using the CRISPR diagnostics‐based SHERLOCK (Specific High Sensitivity Enzymatic Reporter UnLOCKing) technique. RNA fragments of the SARS‐CoV‐2 virus help detect target sequences of approximately 100 copies. The experiment is performed by isothermal amplification of the extracted nucleic acid of samples from patients and then amplification of the viral RNA sequence via Cas13 and is finally read out by a paper dipstick in less than 1 hour. 103 , 104 Huang et al . 105 established a CRISPR‐based assay by a custom CRISPR Cas12a/gRNA complex. They used a fluorescent probe to identify target amplicons produced by standard RT‐PCR or isothermal recombinase polymerase amplification. This method showed specific detection at places not equipped with the PCR systems needed for qPCR diagnostic tests in real time. The analysis allows the identification of SARS‐CoV‐2 positive samples with a test‐to‐response time of approximately 50 minutes and a detection limit of two copies of each sample to be detected. The findings of the CRISPR test on nasal samples collected from persons with COVID‐19 were comparable with matched data achieved from the CDC‐approved RT‐qPCR test. 105
Broughton et al . 106 described the development of a fast (< 40 min), simple‐to‐implement and precise CRISPR‐Cas12‐based lateral flow test to diagnose SARS‐CoV‐2 from RNA extract from a nasal swab. Using artificial reference samples and clinical specimens from patients, comprising patients diagnosed with COVID‐19 disease and 42 patients with other respiratory illnesses, they confirmed their process. This CRISPR‐based approach has provided a visual and quicker alternative option to the SARS‐CoV‐2 real‐time RT‐PCR method used in the US Centers for Disease Control and Prevention, with approximately 100% negative predictive agreement and 95% positive predictive agreement. 106
2.6. LAMP‐based technique
Loop‐mediated isothermal amplification (LAMP) is a new isothermal nucleic acid amplification method with great efficiency. This is used to amplify RNAs and DNAs with high specificity and sensitivity as a result of its exponential amplification feature and six particular target sequences diagnosed by four separate primers. 107 The LAMP assay is rapid and does not need high‐priced reagents or equipment. Furthermore, the gel electrophoresis method is widely utilized for investigation of the amplified items to detect endpoints. Hence, the LAMP test will help to decrease the cost of coronavirus detection. Several strategies for the detection of coronavirus based on LAMP are defined here, as developed and performed in clinical diagnosis. 108
Poon et al . 109 have reported a simple LAMP test in the SARS study and demonstrated the feasibility of this method for SARS‐CoV detection. The SARS‐CoV ORF1b site was selected for SARS detection and amplified in the presence of six primers via the LAMP reaction, and then the amplified products were assessed by gel electrophoresis. The sensitivity and detection levels in LAMP test for SARS are close to those of traditional PCR‐based techniques. Pyrc et al . 110 effectively applied LAMP to HCoV‐NL63 detection with a desirable sensitivity and specificity in mobile cell cultures and clinical specimens. Particularly, one replica of RNA template was found to be responsible for the detection restriction. Amplification is observed as fluorescent dye or magnesium pyrophosphate precipitation. These techniques can be achieved in real time by monitoring the turbidity of the pyrophosphate or fluorescence, which correctly overcome the restriction of endpoint detection. 110
Shirato et al . 111 developed a beneficial RT‐LAMP assay for the diagnosis and epidemiological monitoring of human MERSCoV. This method was highly specific, without any cross‐reaction with other specific respiratory viruses, and detected as few as 3.4 copies of RNA. 111 Subsequently, they developed the RT‐LAMP assay by revealing a sign using a quenching probe (QProbe), which has the same efficiency as the usual real‐time RT‐PCR test with respect to MERSCoV detection. 112
Based on other evidence, a nucleic acid visualization method was developed that combines RT‐LAMP and a vertical flow visualization strip for MERS detection. 113
2.7. Penn RAMP technology
Based on the effectiveness reported by Zhang et al . 104 using the comparatively less sensitive LAMP, the improved sensitivity of the Penn‐RAMP technique achieved by Huang et al . 114 , which is attributable to an updated two‐step LAMP protocol, can prove to be substantially effective as a diagnostic. To amplify specific targets by recombinase polymerase amplification, in which all targets are simultaneously amplified, the Penn‐RAMP requires a preliminary reaction with outer LAMP primers. A next highly precise LAMP reaction is then triggered. Especially, the first stage uses F3 and B3 outer LAMP primers, whereas the other four RAMP primers are further mixed in the stage 2. Compared to normal LAMP, this ‘nested’ concept considerably improved the sensitivity of LAMP by approximately 10–100 times, especially when working with distilled and crude samples. 115 Additionally, when extended to mock trials, the Penn‐RAMP methodology was given a 100% approval rating at 7–10 copies of viral RNA per reaction, compared to a 100% approval rating at the 700 viral RNA copies needed for PCR analysis. 114 , 115
2.8. Droplet digital PCR
For the direct identification and quantification of DNA and RNA targets, droplet digital PCR (ddPCR) comprises an extremely sensitive technique. 116 It has been widely used for infectious disease conditions, particularly because of its ability to identify a few copies of viral genomes accurately and efficiently. 117 If low‐level and/or residual viral existence identification is appropriate, ddPCR quantitative data are much more insightful than those provided by regular RT‐PCR tests. In view of the need to restrict (as far as possible) false‐negative results in COVID‐19 diagnosis, use of the ddPCR can provide a vital support. Even so, the ddPCR assay is still very rarely studied in clinical settings and there is currently no available evidence for European cases. 118
2.9. Next‐generation sequencing (NGS)‐based technique
RNA viruses come in great assortment of varieties, and they are the etiological specialists of numerous significant human and animal infectious diseases. 119
RNA viruses comprise the major variety and are the etiologic agents of very infectious diseases in humans and animals such as SARS, hepatitis, influenza and IB (avian infectious bronchitis). High‐throughput NGS technology has a vital role in primary and accurate diagnosis. 120 In addition, the NGS method can detect whether or not various types of virus comprise a pathogen. The fast novel technique of viruses by NGS, including DNA‐sequencing and RNA‐sequencing has developed the identification of viral diversity. 121 The identification of a huge range of pathogen using NGS technologies is also significant for controlling viral infection caused by a new pathogen. 122 In recent years, the advancement of the NGS method via RNA‐sequencing has enabled us to make great progress in the fast recognition of new RNA viruses. RNA ‐ sequencing detects millions of reversely transcribed DNA fragments from complex RNA samples at the same time using random primers. 123 Chen et al . 122 reported a new duck coronavirus using the RNA‐sequencing method, which differed from that of chicken IBV (infectious bronchitis virus). 122 The new duck‐specific CoV was a possible new species within the genus Gamma‐coronavirus, as shown by sequences of the viral 1b gene from three regions.
In conclusion, the outbreak of a novel virus emerged at the end of December 2019. COVID‐19 spread immediately and challenged medicine, economics and public health worldwide. Numerous evidence proposed that the ACE2 receptors comprise crucial structural proteins for virus budding and entry into host cells. Both transmission from unidentified intermediate hosts to cross‐species and human to human transmission have been recognized. Hence, early detection and isolation of suspected patients can play an essential role in controlling this outbreak. Currently, diagnostic methods for COVID‐19 are numerous; hence, it is imperative to choose a suitable detection protocol. Each of the described techniques has its specific disadvantages and advantages. Both chest CT imaging and RT‐PCR tests are recommended for COVID‐19 patients. However, the use of PCR requires various equipment and a well‐established laboratory. LAMP can be detected with low numbers of DNA or RNA templates within 1 hour. Microarray is an expensive method for COVID‐19 diagnosis, and other newly developed methods also require additional investigation to achieve rapid development and detection in the future. Given that the number of infected cases is rapidly increasing, future studies should reveal the secrets of the molecular pathways of the virus with respect to developing targeted vaccines and antiviral treatments.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
MM and HC drafted the article. MM and HC provided critical revision of the article. SB carried out native editing. NP and AE developed the theory to investigate a specific aspect and supervised the findings of the work, as well as conceived the presented idea. MM, HC, AS, SB, NP and AE discussed the results and contributed to the final manuscript submitted for publication.
Mohamadian M, Chiti H, Shoghli A, Biglari S, Parsamanesh N, Esmaeilzadeh A. COVID‐19: Virology, biology and novel laboratory diagnosis . J Gene Med . 2021; 23 :e3303 10.1002/jgm.3303 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
Malihe Mohamadian and Hossein Chiti contributed equally to this work.
Contributor Information
Negin Parsamanesh, Email: [email protected] .
Abdolreza Esmaeilzadeh, Email: moc.oohay@azer64a .
DATA AVAILABILITY STATEMENT
Log in using your username and password
- Search More Search for this keyword Advanced search
- Latest content
- Publish with us
- About the journal
- Meet the editors
- Specialist reviews
- BMJ Journals More You are viewing from: Google Indexer
You are here
- Volume 1, Issue 1
- Covid-19: virology, variants, and vaccines
- Article Text
- Article info
- Citation Tools
- Rapid Responses
- Article metrics
- Megan Young 1 ,
- Harry Crook 1 ,
- Janet Scott 2 and
- http://orcid.org/0000-0002-6551-2002 Paul Edison 1 , 3
- 1 Faculty of Medicine , Imperial College London , London , UK
- 2 Centre for Virus Research , University of Glasgow , Glasgow , UK
- 3 School of Medicine , Cardiff University , Cardiff , South Glamorgan, Wales , UK
- Correspondence to Dr Paul Edison, Faculty of Medicine, Imperial College London, London W12 0NN, UK; paul.edison{at}imperial.ac.uk
As of 25 January 2022, over 349 million individuals have received a confirmed diagnosis of covid-19, with over 5.59 million confirmed deaths associated with the SARS-CoV-2 virus. The covid-19 pandemic has prompted an extensive global effort to study the molecular evolution of the virus and develop vaccines to prevent its spread. Although rigorous determination of SARS-CoV-2 infectivity remains elusive, owing to the continuous evolution of the virus, steps have been made to understand its genome, structure, and emerging genetic mutations. The SARS-CoV-2 genome is composed of several open reading frames and structural proteins, including the spike protein, which is essential for entry into host cells. As of 25 January 2022, the World Health Organization has reported five variants of concern, two variants of interest, and three variants under monitoring. Additional sublineages have since been identified, and are being monitored. The mutations harboured in these variants confer an increased transmissibility, severity of disease, and escape from neutralising antibodies compared with the primary strain. The current vaccine strategy, including booster doses, provides protection from severe disease. As of 24 January 2022, 33 vaccines have been approved for use in 197 countries. In this review, we discuss the genetics, structure, and transmission methods of SARS-CoV-2 and its variants, highlighting how mutations provide enhanced abilities to spread and inflict disease. This review also outlines the vaccines currently in use around the world, providing evidence for every vaccine's immunogenicity and effectiveness.
Data availability statement
Data are available in a public, open access repository.
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/ .
https://doi.org/10.1136/bmjmed-2021-000040
Statistics from Altmetric.com
Request permissions.
If you wish to reuse any or all of this article please use the link below which will take you to the Copyright Clearance Center’s RightsLink service. You will be able to get a quick price and instant permission to reuse the content in many different ways.
Introduction
Seven coronaviruses can infect humans, all belonging to the alpha or beta subgroups, including 229E (alpha), NL63 (alpha), OC43 (beta), and HKU1 (beta). 1 Over the past two decades, three notable beta coronaviruses (severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002; Middle East respiratory syndrome coronavirus (MERS-CoV) in 2011; and most recently, severe acute respiratory syndrome 2 (SARS-CoV-2) in 2019) have emerged and caused severe illness, resulting in debilitating disease and worldwide deaths. SARS-CoV-2 is the pathogen responsible for the current coronavirus 2019 (covid-19) pandemic and has caused more than 5.59 million deaths in around two years and resulted in multisystem illness in several million people. 2
All viruses change and mutate over time, with most changes having little to no impact. However, some mutations could alter its pathogenic or transmission potential and might, therefore, increase disease severity or hinder the effectiveness of vaccines and therapeutic strategies. The World Health Organization 3 classifies variants of concern as SARS-CoV-2 variants that increase transmissibility, disease severity, or virulence or that decrease the effectiveness of public health measures, diagnostics, therapeutics, or vaccines. Variants of interest are variants with genetic changes predicted to enhance the virulence and transmissibility of the virus, which have been identified to cause community transmission in multiple countries and pose a possible risk to global public health. Lastly, variants under monitoring are those with genetic changes are suspected to affect virus characteristics and have currently unclear phenotypic or epidemiological effects. Variants under monitoring are not typically assigned a name until they are upgraded to variants of interest or concern. The full working definitions of variants of concern, variants of interest, and variants under monitoring can be found on the WHO website for tracking SARS-CoV-2 variants ( www.who.int/en/activities/tracking-SARS-CoV-2-variants/ ). 3 As of 25 January 2022, WHO reports five variants of concern (alpha, beta, gamma, delta, and omicron), two variants of interest (lambda and mu), and three variants under monitoring. 3 Former variants of concern, variants of interest, or variants under monitoring have been reclassified as "formerly monitored variants," owing to these variants no longer circulating, having little impact on the epidemiological situation, or having no concerning properties. 3 Since the beginning of the covid-19 pandemic, the rapid development of effective covid-19 vaccines has taken place around the world. As of 24 January 2022, 33 vaccines have been approved for use in 197 countries, with 10 vaccines having gained emergency use listing approval from WHO. 4
In this review, we provide an overview of the genome and structure of SARS-CoV-2, describing how these elements allow the virus to infect and replicate inside of host cells, before outlining how certain mutations harboured by SARS-CoV-2 variants enhance these abilities. Next, we examine the current state of vaccine development around the world and provide evidence of the effectiveness of booster doses.
Sources and selection criteria
We searched PubMed and Embase databases for covid-19 related articles published between 1 January 2020 and 25 January 2022 and for general coronavirus related articles published from 1 January 2000 onwards. Our search terms included SARS-CoV-2, covid-19, and specific terms including virology, genome, variants, and vaccine. Additional, specific search terms are outlined in online supplemental file 1 . We performed further manual searching for additional articles and data using relevant databases, including who.int, gov.uk, and ecdc.europa.eu/en. Owing to the rapidly evolving nature of the literature involving SARS-CoV-2, we also searched preprint databases including MedRxiv and BioRxiv. We selected studies through different criteria ( online supplemental file 1 ), owing to the various topics discussed here. Overall, studies were selected on the basis of quality and impact factor of publishing journal, with real world studies with large sample sizes of the greatest interest.
Supplemental material
Viral transmission, clinical presentation, and genetic susceptibility of covid-19.
SARS-CoV-2 is predominantly spread via respiratory droplet transmission, spreading between people through close contact, coughing, or sneezing. The virus can also spread through airborne transmission, fomite transmission, and via other modes, such as through biological material including urine and faeces. 5 6 The SARS-CoV-2 virus can survive on surfaces or survive suspended in air droplets for long periods. Indeed, on plastic, stainless steel, and glass surfaces, the half life of the virus is around 5.3, 4.4, and 4.2 hours, respectively, 7 with no difference seen between SARS-CoV-2 variants. 8 Although SARS-CoV-2 can be detected on inanimate surfaces for hours and days, owing to the evaporation of water droplets (the viruses’ living environment), the concentration of the virus plummets rapidly. 9 Protective measures, including use of personal protective equipment, maintenance of indoor ventilation, and disinfection hands and surfaces, can effectively limit the spread of SARS-CoV-2. 10
Once inside the airways, SARS-CoV-2 can infect ciliated, mucus secreting, and club cells of bronchial epithelium, type 1 pneumocytes within the lungs, and the conjunctival mucosa. 11 The clinical presentation of covid-19 is non-specific and heterogeneous, and infection can result in a wide spectrum of symptoms. After an incubation period of 4-14 days, symptoms range from mild to severe disease and, in some instances, can result in death. 12 The most common covid-19 symptoms include fever, cough, dyspnoea, and fatigue, 13 14 while myalgia, gastrointestinal issues, cognitive deficits, and other symptoms are reported. Asymptomatic individuals can also test positive for covid-19. 15 16 Although the entire population is susceptible to covid-19 infection, some subgroups within the general population are more susceptible to developing poorer clinical outcomes.
Risk factors associated with increased probability of hospital admission, severe disease, and fatal outcome with covid-19 have been identified. Older age 17–19 ; male sex 20 21 ; belonging to an ethnic minority group 21 22 ; and comorbidities including diabetes, hypertension, and lung disease, 18 23–25 malignancy, and immunodeficiency 26–28 have all been associated with more severe covid-19. The duration and treatment of covid-19 symptoms will also have profound influences on the severity of disease and the acute and long term outcomes after recovery. The host genetic background is thought to have an influence on the susceptibility and severity of covid-19, possibly explaining the broad spectrum of clinical manifestations that can develop in seemingly similar individuals. A meta-analysis, consisting of 49,562 patients with covid-19 across numerous ancestry groups, identified four gene loci associated with susceptibility to covid-19 (SLC6A20, RPL24, ABO, PLEKHA4) and nine associated with increased risk of severe covid-19 (LZTFL1, FOXP4, TMEM65, OAS1, KANSL1, TAC4, DPP9, RAVER1, and IFNAR2). 29 Meanwhile, genome wide association studies spanning across Europe, the US, and the UK identified a gene cluster on chromosome three (chr3p21.31) as being strongly linked with susceptibility and severity of covid-19. 30 31 Polymorphisms in the genes of the angiotensin converting enzyme 2 (ACE2) receptor and transmembrane protease serine 2 (TMPRSS2) have also been shown to enhance SARS-CoV-2 viral entry, 32 33 with differential polymorphisms seen across ethnic minority populations, which might partly explain why certain ethnic groups are more susceptible to severe covid-19. Increased ACE2 receptor levels have also been associated with other risk factors of covid-19, including smoking and increasing age. 34 The use of polygenetic risk scores might be useful in determining an individual’s risk for developing severe disease caused by covid-19. 35 A polygenetic risk score infers a person’s risk of susceptibility to, or development of, a certain disease based on the total number of genomic variations they possess. Determining polygenetic risk scores with the inclusion of comorbidities, such as chronic obstructive pulmonary disease, 36 or other aspects such as coagulation factors, 37 could improve the usefulness of these scores in determining a person’s risk of severe covid-19.
Virology of SARS-CoV-2
SARS-CoV-2 is a positive stranded RNA virus belonging to Coronaviridae family. Coronaviruses, which have crown-like appearances, are the largest known RNA viruses and are thought to primarily infect vertebrates. 38 39 SARS-CoV-2 belongs to the beta genus of the coronaviruses and has a genome size varying from 29.8 to 29.9 kb. 40 Human coronavirus genomes consist of a variable number of open reading frames (ORFs). Following the typical 5’ to 3’ order, the beginning two thirds of the SARS-CoV-2 genome contains two ORFs (ORF1a and ORF1b) that, inside the host cell, are translated at the rough endoplasmic reticulum into polyprotein 1a (pp1a) and polyprotein 1ab (pp1ab), respectively. 40 These polyproteins are cleaved into 16 non-structural proteins (nsp): nsp1-11, from pp1a; and nsp12-16, from pp1ab. The proteolytic release of nsp1 occurs rapidly, which enables it to interfere with translation processes of the host cell by inducing cellular mRNA degradation. 41–43 Nsp2-16 contain the viruses’ replication and transcription complex and encode multiple enzymes with many functions, including proteases, helicase, polymerase, exonuclease and endonuclease, N7-methyltransferase and 2’O-methyltransferase, and de-ubiquitination enzymes. 44 45
The final third of human coronavirus genomes contain genes that encode structural and accessory proteins. The four major structural proteins encoded here are the nucleocapsid (N), membrane (M), envelope (E), and spike glycoprotein (S) proteins. 46 47 The N protein is associated with the viral RNA genome, is involved in RNA synthesis regulation, and interacts with the M protein during viral budding. 39 48 The M protein is important for viral assembly, it contains a short N-terminal domain that projects onto the external surface of the envelope and a long internal C-terminal. 39 The E protein function is largely unknown; however, along with the N and M proteins, it is required for viral assembly and release. 47 Lastly, the S protein gives coronaviruses their characteristic spikes that compose their crownlike appearance. This protein projects through the viral envelope, is heavily glycosylated, and regulates host cell membrane receptor binding and fusion of the viral and cellular membrane. 49 The functions of the 11 accessory proteins encoded within the one-third closest to the 3’ end of the SARS-CoV-2 genome are not fully understood. These accessory proteins are encoded by the ORF3a, ORF3b, ORF3c, ORF3d, ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORC9c, and ORF10 genes. Some of these proteins, including ORF3b, ORF6, ORF7a, and ORF8, are interferon antagonists that impair the host cell immune response, 50–53 whereas ORF3a might promote virus release 54 and is involved in apoptosis of host cells through caspase-3 activation. 55 ORF9b and ORF9c are known to suppress the host antiviral response by interacting with host cell organelles, 56–58 whereas a clear understanding of the functions of ORF3c, ORF7b, and ORF10 remains unclear. 59 Figure 1 (A,B) depicts the genome and structure of SARS-CoV-2.
- Download figure
- Open in new tab
- Download powerpoint
Genome and structure of SARS-CoV-2. (A) SARS-CoV-2 genome and spike (S) protein amino acid composition. The SARS-CoV-2 genome is about 30 000 base pairs (bp) long and consists of open reading frames (ORF) and elements that are essential for the virus’ structure. The S protein is responsible for binding and entry into host cells. SARS-CoV-2 variants of concern contain various S protein non-synonymous mutations that result in amino acid changes in the receptor binding domain (orange bracketed text) and the S1/S2 subunit interface (black bracketed text), which have been shown to enhance transmissibility of the virus. Variants of concern include alpha (α), beta (β), gamma (γ), delta (δ), and omicron (O). (B) SARS-CoV-2 structure. SARS-CoV-2 is an RNA virus that has a crown-like appearance and contains four major structural proteins: nucleocapsid (N), spike (S), envelope (E), and membrane (M). (C) Viral S protein and human angiotensin converting enzyme 2 (ACE2) interaction. The SARS-CoV-2 S protein directly interacts with human ACE2 receptors in order to gain entry into host cells. The receptor binding domain (RBD) of the S protein tightly binds to ACE2. (D) S protein structure. The S protein protrudes out from the main SARS-CoV-2 bulk and is comprised of two subunits: S1 and S2. S1 contains the RBD, which directly interacts with the human ACE2 receptor, while the S1/S2 interface contains a furin cleavage site that is cleaved to allow S2 to fuse with the host cell membrane. Both the RBD and the S1/S2 interface contain transmissibility increasing mutations that are harboured in variants of concern
The S glycoprotein is composed of two functionally distinct subunits (S1 and S2) and is essential for viral entry into host cells. The N-terminal S1 domain of the protein contains the receptor binding domain (RBD) that directly interacts with the ACE2 receptor on the host cell, which is the primary receptor that SARS-Cov-2 uses for cell entry. 60 The C-terminal S2 domain fuses the host and viral membranes to allow for entry of the viral genome into the host cell. 61 The subunits of the trimeric S complex are either in a closed (pre-fusion stage) or open (post-fusion stage) conformation, 62 with one subunit always in an open conformation to allow for ACE2 recognition and binding. 63 The RBD itself consists of five anti-parallel β strands surrounded by several α helices. 64 From closed to open conformation, the RBD undergoes structural rearrangement whereby the globular head region rotates clockwise, which alters is elecropotential surface. 64 Once positioned, numerous residues within the RBD form either hydrogen bonds or salt bridges with residues of the ACE2 receptor, allowing for tight binding, 65 while the concave structure of the RBD allows for three distinct binding regions. 64 Following binding between the S protein and the host cell receptor, host cell proteases cleave the S protein, causing the release of the S2 domain which allows for fusion and cell entry. 66 Figure 1 (C,D) shows the structure and function of the S protein.
The ACE2 receptor is expressed in numerous cell types throughout the human body, including in the lungs, oral and nasal mucosas, heart, gastrointestinal tract, kidneys, liver, spleen, and brain, 67 highlighting the widespread infection that SARS-CoV-2 can inflict. Meanwhile, TMPRSS2, a host cell protease, facilitates fusion of the viral and host cell membranes, 68 and could have a role in the spread of the virus in the airways. 68 Host cell cathepsin L might also aid in SARS-CoV-2 cell entry by cleaving the S protein. 69 Indeed, a clinically approved protease inhibitor has been shown to block SARS-CoV-2 cell entry. 70 Figure 2 depicts the mechanism by which SARS-CoV-2 gains entry into and replicates inside host cells, and summarises the host cell immune response.
Viral entry and host response. (A) At the alveolar epithelial cell layer. Epithelial cells in the lungs express both angiotensin converting enzyme 2 (ACE2) receptors and transmembrane protease serine 2 (TMPRSS2), allowing for infection by SARS-CoV-2. Replication of the virus within these cells induces an intense immune response that attracts monocytes, T cells, and macrophages and, in some instances, can result in a cytokine storm. (B) Within nearby blood vessels. Cytokines produced by the epithelial cell layer are released into blood vessels supplying the infected tissue, which causes the recruitment of further immune cells to the area, driving the damaging inflammatory response further. Circulating cytokines also create a systemic inflammatory environment. (C) Adaptive immune response. Circulating lymphocytes carry viral antigens to lymph nodes and bone marrow to begin the adaptive immune system processes whereby B cells, and later antibodies, are activated. (D) SARS-CoV2 host replication. The SARS-CoV-2 virus uses the ACE2 receptor and TMPRSS2 to gain entry into human cells. Following release of the viral RNA within the host cell, the virus uses the host endoplasmic reticulum (ER) and Golgi apparatus to produce and manufacture new viral particles, which are released out of the cell to infect other cells and new hosts
Variants of SARS-CoV-2
Most viral mutations have a limited impact on the viruses’ ability to infect, replicate, escape host immunity, and transmit; however, certain mutations can give a viral strain a competitive advantage and, through natural selection, give it the ability to become dominant. Many mutations observed in SARS-CoV-2 variants are found within the RBD or the N-terminal domain of the S protein, which alters the three dimensional structure of the S protein. Not only can these changes affect the transmission abilities of the virus, but it can also allow it to better escape the immune response, such as from neutralising antibodies either elicited through vaccine administration or natural infection.
The SARS-CoV-2 virus has mutated numerous times, with estimates suggesting that circulating lineages acquire nucleotide mutations at rates of around one to two mutations per month. 71 The current method of identifying variants relies on the use of genomic testing such as whole genome sequencing, partial S gene sequencing, or assays based on nucleic acid amplification. 72 The aspects of different variants that most people experience, however, is the clinical symptoms they inflict. Certain variants (eg, alpha, delta) induce a greater risk of severe disease and death, 73 while others (eg, omicron) are more likely to induce milder symptom. 74 75 Moreover, individual symptoms can differ between variants. For example, the gamma variant is associated inflicting anosmia and dysgeusia, 76 which is less commonly seen in omicron infections. Moving forward, the clinical themes and symptoms associated with emerging variants should be elucidated rapidly so that the public and healthcare professionals can rapidly identify possible cases of covid-19.
WHO has tracked and monitored SARS-CoV-2 variants since the covid-19 pandemic began to identify variants of concern. As of 25 January 2022, WHO reported five variants of concern, two variants of interest, and three variants under monitoring ( table 1 ). 3 Here, we report studies that compare SARS-CoV-2 variants to the primary virus strain. The primary strain is the strain of the virus that first emerged in Wuhan, China at the end of 2019 and spread around the world in the first wave of infections, which is often also referred to as the Wuhan-Hu-1, B.1, or wildtype strain.
- View inline
SARS-CoV-2 variants and related spike protein mutations
Variants of concern
Alpha variant b.1.1.7.
The alpha SARS-CoV-2 variant of the B.1.1.7 lineage was first documented in the UK in September 2020 and classified as a variant of concern on 18 December 2020. 3 77 This variant contains S protein mutations that have potential biological effects. Firstly, the S protein residue 501, a key contact residue within the RBD, forms a portion of the binding loop in the contact region of the ACE2 receptor, forms a hydrogen bond with the Y41 residue of the ACE2 receptor, and stabilises the ACE2 K353 residue. 65 78 79 The alpha variant has an N501Y mutation, which increases the binding affinity of the RBD to the ACE2 receptor. 80 Next, the P681H mutation contained within the alpha variant is located immediately adjacent to the 682-685 furin cleavage site, at the interface of the S1 and S2 domains. 81 The S1/S2 furin cleavage site prompts entry into respiratory epithelial cells and partly determines the transmissibility of the virus, 82–84 while the P681H mutation makes the furin cleavage site less acidic, meaning it is more effectively recognised and cleaved. 85 86 Alpha also contains a D614G mutation, located within the S1/S2 furin cleavage site, which increases SARS-CoV-2 binding affinity to the ACE2 receptor and increases infectivity. 87 Other mutations within the alpha variant enhance the ability of the virus to escape antibody detection, such as the two amino acid deletion at sites 69-70 in the N-terminal domain of the S protein, 88 89 while other mutations show limited or no effects. 90 In February 2021, viruses of the B.1.1.7 lineage with the added S protein mutation E484K were identified, which could have threatened vaccine effectiveness owing to the mutation conferring an increased resistance to neutralising vaccine elicited and monoclonal antibodies. 91 This mutation had limited effects, however, and variants containing it failed to dominate.
Epidemiological studies have explored the alpha variant, with a case-control study of 27 633 respiratory samples originating from 20 primary care centres in Madrid, Spain, finding that the probability of admission to an intensive care unit was twice as high in patients infected with the alpha variant compared with those infected with the primary strain. 92 Furthermore, this variant became the dominant strain within four months, and led to an increase in disease burden as a result. 92
Meanwhile in Cannes, France, infection with the alpha variant was associated with a 3.8-fold higher risk of transfer to intensive care or death compared with the primary strain, as determined through a retrospective cohort study of 158 patients with covid-19. 93 A large retrospective cohort study including a total of 476 973 participants found that, during the third covid-19 wave in Canada, where 91% of infections were caused by the alpha variant, the risk of both hospital admission (adjusted odds ratio 1.57) and death (1.52) was higher than primary strain infections. 94 Overall, the alpha variant was about 50-70% more transmissible and was associated with a 30-60% increased risk of hospital admission and death compared with the primary strain. 95–100
The alpha variant was found to have a minimal impact on the effectiveness of current vaccines, 101 102 while the risk of reinfection remained similar for this variant as with previous ones. 103 On 3 September 2021, the European Centre for Disease Prevention and Control (ECDC) reclassified the alpha, and the alpha +E484K mutation variants from a variant of concern to a de-escalated variant. 104
Beta variant B.1.351
The beta SARS-CoV-2 variant, of the B.1.351 lineage, was first documented in South Africa in May 2020. 3 This variant contains five S protein mutations of interest: N501Y, E484K, D614G, K417N, and A701V. Like the alpha variant, the beta variant contains the mutations N501Y, E484K, and D614G, which increase ACE2 receptor binding affinity, 80 87 increase virulence, 105 and enhance resistance to neutralising antibodies. 91 106 The K417 residue of the SARS-CoV-2 S protein interacts with the D30 residue of the ACE2 receptor, forming a salt bridge across the central contact region, 65 78 although the K417N mutation appears to have a limited impact on ACE2 receptor binding. 80 The A701V mutation is located close to the furin cleavage site but has a minimal impact on transmissibility or antibody resistance. 101
In a genomic and epidemiological study, researchers concluded that the beta SARS-CoV-2 variant had a selective advantage over previous variants from its increased transmissibility and immune escape abilities, 107 108 whereas the E484K/N501K mutations enhanced the binding affinity of the beta variant and, hence, increased its transmissibility. 109 A retrospective cohort study of 22 068 participants found that infection with the beta variant was associated with an increased risk of hospital admission compared with an infection with a non-variant of concern (hazard ratio 2.30). 100 Overall, the beta variant is about 25-50% more transmissible, is associated with a possible increase in risk of hospital mortality, and has enhanced resistance to antibody neutralisation compared with previous variants. 107 108 110
Gamma variant P.1
The gamma variant is of the P.1 lineage and was first reported in November 2020 from travellers returning to Japan from Brazil, and was later discovered in Brazil. 3 111 This variant contains the following S protein mutations of interest: K417T, E484K, N501Y, D614G, and H655Y. 104 As mentioned, the N501Y and D614G mutations increase both ACE2 receptor binding affinity and infectivity of the virus. 80 87 The N501Y, K417N/T, and E484K mutation trinity, meanwhile, is shared by both gamma and beta variants, and is associated with enhanced infectivity and lethality compared with the N501Y mutation alone, possibly from tighter binding of the S protein to the ACE2 receptor due to increased electrostatic contribution. 112 The gamma variant also includes the H655Y mutation, which was found to provide enhanced viral escape abilities from multiple human monoclonal antibodies in vitro. 113
The gamma variant is associated with heightened transmissibility, 109 110 114 with one study concluding that it possesses a 1.7-fold to 2.4-fold increased transmissibility compared with previous variants. 115 Additionally, the wave of infections caused by the gamma variant in Brazil was associated with a 13% increase in death rate compared with the previous wave, suggesting the greater virulence held by the gamma variant than by previous viral strains. 116
A surveillance study from seven European countries concluded that the gamma variant was associated with a higher risk of admission to hospital (adjusted odds ratio 2.6) and intensive care (2.2) when compared with cases of non-variants of concern. 117 In Manaus, Brazil, the resurgence of covid-19, despite high seroprevalence, suggested that the gamma variant had a moderate resistance to neutralising antibodies, 118 however, the variant has been shown to be significantly less resistant to neutralising antibodies than other variants, including the beta variant. 119
Delta variant B.1.617.2
The delta variant, from the B.1.617.2 lineage, was first documented in India in October 2020 and was classified as a variant of concern on 11 May 2021. 3 The S protein mutations of interest P681R and D614G are also located in the delta variant 104 and similarly affect its ACE2 receptor binding affinity and transmissibility. 106 120 121 Unlike the E484K mutation seen in previous variants, the delta variant contains the E484Q mutation that, along with a L452R mutation also located within the RBD, causes significantly higher affinity for the ACE2 receptor than the primary strain or the E484K mutation alone. 122 The L452R mutation alone results in greater RBD-ACE2 receptor binding affinity and enhanced escape from neutralising antibodies. 123 124 Lastly, the delta variant contains the T478K mutation, located on the interface between the S protein and the ACE2 receptor when bound, which increases the electrostatic potential of the S protein and enhances binding affinity. 125
The delta variant quickly became the dominant variant in the UK, 126 US, 127 Europe, and around the world. 128 The mutations present in the delta variant enhanced the transmissibility of the virus as a result of increased binding affinity to the ACE2 receptor. 109 The reproduction number of the delta variant is estimated to be 97% greater than that of non-variants of concern or non-variants of interest, and about three times that of the alpha, beta, and gamma variants. 110 This increased reproductivity highlights the delta variant's competitive advantage over earlier ones and how it rapidly became the dominant strain globally. The fast replication rate of delta probably contributes to its increased transmissibility compared with the alpha, beta, and gamma variants. In a cohort study consisting of 167 infections, the delta variant could be detected by polymerase chain reaction within the first four days from exposure, whereas non-delta covid-19 infections could be detected after only six days. 129 Furthermore, people infected with the delta variant were found to have significantly higher viral loads than people infected with other strains, 129 including the beta variant. 130 The delta variant is also thought to better escape neutralisation, with the frequency of post-vaccination infections much higher for the delta variant than infections with the primary strain in India, 131 and blood serum samples from individuals who had received one dose of a covid-19 vaccine showing minimal neutralisation of the delta variant. 132
The delta variant is also associated with an increased disease severity. In Scotland, infection with the delta variant was associated with an increased risk of hospital admission (hazard ratio 1.85) compared with infection with the alpha variant. 133 Compared with infections involving non-variants of concern, North American retrospective cohort studies showed that infection with the delta variant was associated with a 108% 134 or hazard ratio of 2.28 (95% confidence interval 1.56-3.34) 100 increased risk of hospital admission, a 234% increased risk for admission to intensive care, and a 132% increased risk of death. 134 Lastly, in a cross sectional study of 6238 individuals infected with the delta variant and 3262 infected with the primary strain in India, researchers found that the risk of death was around 1.8 times higher for delta infections, while the delta variant also infected and induced symptoms in a greater proportion of younger people (age 0-19 years) than did the primary strain. 131
Omicron variant B.1.1.529
The omicron variant is of the B.1.1.529 lineage and was first discovered in November 2021 in South Africa and Botswana before being detected in multiple countries and classified as a variant of concern on 26 November 2021. 3 This variant contains over 30 S protein mutations, 104 23 of which have been previously identified, including K417N, T478K, E484A, D614G, H655Y, P681H, and N501Y. 135 Fifteen omicron mutations are contained within the RBD, 17 providing the variant with a substantially enhanced binding affinity to the ACE2 receptor. 135 136 In addition, various single mutations in the RBD of the omicron variant impair the effectiveness of neutralising antibodies, including K417N, N440K, G446S, E484A, Q493K, G496S, G339D, S371L, and S375F. 17
The emergence of omicron has been followed by a surge of infections worldwide. Early data from South Africa have indicated that the proportion of covid-19 infections caused by the omicron variant rose from 3% in early October 2021 to 98% by early December 2021. 137 In late December 2021, meanwhile, the doubling time for the number of omicron infections was between two and three in the UK, US, and much of Europe, 138 139 highlighting the transmissibility of this variant. The mutations in the omicron variant that enhance its binding affinity 135 136 and ability to escape neutralising antibodies 17 probably drove its rapid spread, as did its fast replication rate, which is around 70 times faster than the delta and primary strains. 140 The reinfection rate of the omicron variant has also been found to be more than ten times higher than that of previous variants in studies from Scotland 141 and South Africa. 142
The omicron variant has extensive but incomplete escape abilities from naturally acquired and vaccine induced immunity. 143 144 Compared with the delta variant, the omicron variant needs around a 10-fold increased antibody titre to be neutralised, after vaccination with either the ChAdOx1 nCoV-19 (Oxford-AstraZeneca) or BNT162b2 (Pfizer-BioNTech) vaccines. 145 Indeed, blood serum from individuals who had received two doses of the BNT162b2 vaccine showed more than a 25-fold reduction in neutralising antibody titres against the omicron variant compared with the primary strain. 146 T cell responses to the omicron variant could remain intact, however. Data from one preprint study indicated that 70-80% of the T cell response targeting the S protein was maintained in those individuals vaccinated or with previous infection, while the magnitude of T cells cross reacting with the omicron variant was similar to that of both delta and beta variants. 147 Furthermore, data from Pfizer-BioNtech revealed that 80% of the epitopes in the omicron variant S protein that are recognised by CD8 T cells were not affected by the variant’s mutations, after two doses of the vaccine. 146 T cell responses induced from vaccination or prior infection could, therefore, provide some protection from severe disease.
Recent real world evidence has implied that omicron infection is milder in severity than previous variants. In an early South African analysis, the risk of hospital admission (adjusted odds ratio 0.2) was lower for omicron infections than for non-omicron infections, 137 while omicron infected individuals had a lower risk of severe disease than delta infected individuals (0.3). 137 In December 2021 in England, omicron infections were found to induce a greatly reduced risk of hospital admission or presentation for emergency care than delta infections. 74 75 The decreased disease severity inflicted by the omicron variant could be due to its reduced capacity for replication in lung tissue, which was found to be more than 10 times less in lung tissue than the delta variant. 140 Concordantly, the S protein of the omicron variant is less efficient at cleaving the ACE2 receptor and entering cells of lung organoids, 145 and is also less able to cause fusion between lung cells than the S protein of the delta variant, 145 which is often observed in severe covid-19. The reduction in replication within the lungs, and the preservation of T cell responses probably contribute to the milder disease exerted by the omicron variant.
The original Omicron variant is referred to as BA.1, due to the detection of several sublineages of the variant in circulation. While the emergence of BA.1 coincided with a wave of covid-19 infections around the world due to its higher transmissibility and increased risk of reinfection than previous variants, 148 sublineages BA.2 and BA.3 are also circulating, with BA.2 now responsible for an increasing number of the reported cases. 149 The current data remains limited, however, the UK Health Security Agency report that BA.2 has an increased growth rate compared to BA.1 although this report did not find any evidence of a difference in vaccine effectiveness between the two sublineages of the Omicron variant. 150 Indeed, the REACT-1 study of covid-19 transmission concluded that BA.2 had a daily growth rate additive advantage of 0.4 compared to BA.1. 151 The risk of hospitalisation does not seem to be higher for BA.2 infection in comparison to BA.1, however. 152 Emerging sublineages of the Omicron variant will be required to be monitored and reported upon for the foreseeable the future.
Although the omicron variant seems to manifest in mild disease, high infection numbers could still result in high rates of hospital admission and death in those individuals vulnerable to the virus. Omicron case numbers could be beginning to peak, however. In South Africa, a 29.7% decrease in weekly covid-19 infections were reported in the week ending 25 December 2021, compared with the previous week, and the omicron wave is said to have passed. 153 Concerningly, global case numbers continue to rise rapidly 154 and many countries will continue to feel the pressure exerted by the wave of omicron infections.
Variants of interest
Lambda variant c.37.
The lambda variant, of the C.37 lineage, was first documented in Peru in December 2020 and was designated as a variant of interest on 14 June 2021. 3 This variant contains the S protein mutations D614G, L452Q, and F490S. 104 The L452Q mutation, located within the RBD, enhances binding affinity to the ACE2 receptor and increases the infectivity of the lambda variant, 155 while, together L452Q and F490S, increasing the variant's resistance to vaccine elicited antibody neutralisation. 155 Furthermore, F490S was identified as being a high risk mutation for enhancing abilities to escape neutralisation. 155
Infectivity of the lambda variant could be higher than that of the alpha, gamma, and other D614G containing variants, 156 suggesting that lambda could spread more rapidly and effectively. Additionally, compared with the primary SARS-CoV-2 virus, antibody neutralisation was found to decrease by 3.05-fold for the lambda variant, higher than that for the gamma (2.33-fold) and alpha (2.03-fold) variants. 156 However, findings from a preprint study suggest that the lambda variant can be neutralised by monoclonal antibodies, and that current vaccines are protective against this variant. 155
Mu variant B.1.621
The mu variant, from the B.1.621 lineage, was first documented in Columbia in January 2021 before receiving designation as a variant of interest on 30 August 2021. 3 This variant contains the S protein mutations E484K, N501Y, D614G, and P681H. 104 Mu also contains the S protein mutation R346K, located within the RBD, 104 157 which can induce large, binding, free energy changes that disrupt the binding of antibodies to the S protein and enhance the ability of the variant to escape neutralisation. 158 As discussed, the E484K, N501Y, D614G, and P681H mutations have been shown to increase transmissibility 80 85 87 105 109 112 120 121 and neutralisation escape, 91 106 suggesting that the mu variant is likely to be more infectious than the primary strain.
Although the lambda and mu variants have been outcompeted by the delta and now omicron variants, the development and spread of these variants of interest will need to be closely monitored and studied to appreciate their pathogenicity, transmissibility, and virulence.
Variants under monitoring
As of 25 January 2022, three variants under monitoring were listed by WHO 3 ( table 1 ).
The covid-19 pandemic prompted a rapid international search for safe and effective vaccines against the SARS-CoV-2 virus. In line with previous vaccine development, including for both SARS-CoV and MERS-CoV, the S protein was a key target for covid-19 vaccine development. 159 As of 24 January 2022, 33 approved vaccines are in use in 197 countries, with 10 vaccines approved for emergency use by WHO ( online supplemental table ). 4 , 115 133 160–251 As of 25 January 2022, 194 vaccines were in pre-clinical development and 140 were in clinical development. 252 Numerous studies have explored the effectiveness of approved vaccines; however, large variations in vaccine effectiveness are reported. This variability is probably due to several factors in the studies, including the country, date, and population size of the study, as well as the SARS-CoV-2 variants circulating during the study period. These factors, along with how the effectiveness is reported, mean that it is difficult to compare vaccines and fully understand how effective each vaccine is. Here, we review the covid-19 vaccines in use around the world.
BNT162b2 (Pfizer-BioNtech)
The BNT162b2 vaccine (Comirnaty) is a lipid nanoparticle formulated, nucleoside modified, mRNA vaccine encoding a modified SARS-CoV-2 S protein that was developed through a collaborative effort between Pfizer (New York, NY, USA) and BioNTech (Mainz, Germany). 62 160 The vaccine was listed by WHO for emergency use on 31 December 2020 253 and, as of 24 January 2022, has been approved for use in 136 countries. 4
Following BNT162b2 vaccination, a response based on T helper 1 (Th1) cells is observed along with elevated levels of tumour necrosis factor α, interferon gamma, and interleukin 2, compared with placebo. 254 255 The highest neutralisation titres are found between seven and 14 days after the second dose, 256 while those individuals previously infected with covid-19 showed a fourfold increase in antibody binding and an 18-fold increase in neutralisation titres compared with previously uninfected individuals after two vaccine doses. 257 The BNT162b vaccine is well tolerated, with limited reactogenicity. Redness and swelling at injection site have been reported, although mild or moderate pain at the injection site is the most commonly reported reaction to vaccination. 256 Fatigue, muscle pain, headache, and chills are other commonly reported symptoms after BNT162b2 vaccination. 258 The rate of systemic reactions after a second dose of BNT162b has been found to be 1.7 to two times higher than after a first dose, possibly suggesting an immunity boosting effect. 259 Many safety reports of this vaccine describe no serious adverse events, 256 259 260 but a large study of 884 828 pairs of individuals, split 1:1 based on vaccination status, found that BNT162b2 was associated with an increased risk of myocarditis, lymphadenopathy, appendicitis, and herpes zoster infection. 261 Although rare, allergic reactions or anaphylaxis has also been reported after BNT162b2 vaccination. 258 The online supplemental table outlines clinical trial and real world data for vaccine effectiveness. 115 133 160–251
ChAdOx1 nCoV-19 (Oxford-AstraZeneca)
The ChAdOx1 nCoV-19 vaccine (AZD1222, Vaxzevria) is a non-replicating vector of the chimpanzee adenovirus ChAdOx1, modified to encode the SARS-CoV-2 S protein. 262 Developed through collaboration between the University of Oxford and AstraZeneca (Cambridge, UK), this vaccine was listed by WHO for emergency use on 16 February 2021, 253 and has been approved for use in 137 countries, as of 24 January 2022. 4 WHO has granted emergency use listing to two versions of this vaccine (AZD1222 and Covishield) in order to use Covishield as part of their worldwide COVAX initiative, which is being produced by the Serum Institute of India and AstraZeneca-SKBio (Republic of Korea). 263
Following ChAdOx1 nCoV-19 vaccination, substantial antibody production (predominantly of IgG1 and IgG3 subclasses) is seen, as well as a Th1 cell response with increased expression of interferon γ and tumour necrosis factor α. 122 264 One dose of the ChAdOx1 nCoV-19 vaccine has been shown to produce a neutralising antibody response in 91% of participants, while a second dose has resulted in 100% of participants producing neutralising antibodies. 265 Mild and moderate itchiness, pain, redness, swelling, tenderness, and warmth are common local reactions, while chills, fatigue, fever, headache, muscle ache, and nausea are commonly reported systemic reactions after vaccination. 265 Rare symptoms, including severe chest pain, nasal bleeding, and allergic reactions have also been reported after vaccination. 266 The online supplemental table outlines clinical trial and real world data for vaccine effectiveness. 115 133 160–251
Ad26.COV.2.S (Johnson & Johnson)
The Ad26.COV.2.S vaccine is a non-replicating adenovirus vector, modified to contain the SARS-CoV-2 S protein in a pre-fusion stabilised conformation and requires only one dose. 161 This vector was developed from the recombinant human adenovirus type 26 by the Janssen pharmaceutical company Johnson & Johnson (New Brunswick, NJ, USA), 161 and was listed by WHO for emergency use on 12 March 2021. 253 As of 24 January 2022, Ad26.COV.2.S has been approved for use in 106 countries. 4
The Ad26.COV.2.S vaccine induces the production of a variety of antibody subclasses, such as immunoglobulins G, M, and A, and promotes several non-neutralising antibody responses, including the activation of CD4 and CD8 Th1 cells and the production of interferon γ, interleukin 2, and tumour necrosis factor α. 267 268 Although neutralising antibody responses induced by the vaccine are reduced against SARS-CoV-2 variants, non-neutralising antibody and T cell responses have been found to be preserved against variants of concern, 267 and a prior covid-19 infection significantly increases levels of S protein binding antibodies, antibody dependent cellular cytotoxicity, and neutralising antibodies against variants of concern (including the beta and delta variants). 269 Ad26.COV.2.S is safe and well tolerated. In a large clinical trial, where 19 630 participants received Ad26.COV2.S and 19 691 received placebo, headache, fatigue, and myalgia were the most common systemic reactions, while pain at the injection site was the most common local reaction after vaccination. 161 Like other vaccines, Ad26.COV.2.S has been associated with serious adverse events, such as allergic reactions and cerebral venous sinus thrombosis; however, these events are rare. 258 270 The online supplemental table outlines clinical trial and real world data for vaccine effectiveness. 115 133 160–251
mRNA-1273 (Moderna)
The mRNA-1273 vaccine (Spikevax) developed by Moderna (MA, USA) is a lipid-nanoparticle encapsulated mRNA vaccine expressing the SARS-CoV-2 S protein that has been pre-fusion stabilised. 162 This vaccine gained WHO approval for emergency use listing on 30 April 2021, 253 and as of 24 January 2022, has been approved for use in 85 countries. 4
The mRNA-1273 vaccine elicits a strong CD4 Th1 cell response, with tumour necrosis factor α, interferon γ, and interleukin 2 expression increased following vaccination, 271–273 while neutralising antibody titres have been shown to increase up to until around 28 days after the second vaccine dose, and remain consistently high after that. 274 Fatigue, muscle pain, headache, chills, joint pain, and pain/reaction at the injection site are common adverse effects caused by the mRNA-1273 vaccine, 162 258 while serious adverse effects are often avoided. 162 274 Serious adverse events, including allergic reaction and anaphylaxis, are rare but not inconceivable after mRNA-1273 vaccination. 258 The online supplemental table outlines clinical trial and real world data for vaccine effectiveness. 115 133 160–251
Other covid-19 vaccines listed by WHO for emergency use
In addition to the covid-19 vaccines described above, five other vaccines have gained emergency use listing by WHO. Firstly, the Sinopharm BBIBP-CorV covid-19 vaccine (Covilo) was developed by the Beijing Bio-Institute of Biological Products, a subsidiary of China National Biotec Group, and was approved by WHO for emergency use on 7 May 2021. 253 This vaccine is made from the SARS-CoV-2, 19nCoV-CDC-Tan-HB02 strain, which is produced in Vero cells, inactivated by β propiolactone, and then purified and absorbed with aluminium hydroxide. 275
Next, the CoronaVac vaccine, developed by Sinovac Biotech (Beijing, China), was listed for WHO emergency use on 1 June 2021. 253 Like the BBIBP-CorV vaccine, this vaccine is a Vero cell based, aluminium hydroxide adjuvanted, beta propiolactone inactivated vaccine, but it is based on the SARS-CoV-2 CZ02 strain. 276 Covaxin (BBV152) is a whole virion inactivated, SARS-CoV-2 vaccine formula developed by Bharat Biotech International (India), 277 which gained approval for emergency use listing from WHO on 3 November 2021. 278
Lastly, Covovax and its originator, Nuvaxovid (NVX-CoV2372), were both developed by Novavax (MD, USA) and the Coalition for Epidemic Preparedness Innovations (Oslo, Norway), and were listed by WHO for emergency use on 17 and 21 December 2021, respectively. 279 280 Both vaccines are manufactured by the same technology, and consist of a recombinant SARS-CoV-2 S protein nanoparticle combined with the adjuvant Matrix-M as a coformulation. 281 These vaccines produce similar immune responses to those already discussed. Studies assessing the efficacy of these vaccines are outlined in the online supplemental table . 115 133 160–251
Other approved covid-19 vaccines
In addition to the vaccines that have received emergency use listing from WHO, vaccines around the world have been developed, tested, and approved to prevent covid-19 infection. As of 24 January 2022, 33 vaccines, including those described above, have been approved in at least one country. 4 The remaining 23 approved vaccines are outlined in table 2 .
Summary of vaccine efficacy across vaccines approved by WHO for emergency use
Waning immunity and boosters
Throughout the covid-19 pandemic, emerging variants have threatened the effectiveness of vaccines ( online supplemental table ). 115 133 160–251 Simultaneously, waning immunity after vaccination questions how long vaccines remain effective and highlights the importance of booster doses. Indeed, protection against SARS-CoV-2 after vaccination decreases over time, both in terms of antibody titres 282–284 and vaccine effectiveness. 163 285–287 However, cellular responses, such as T cell immunity, could persist for longer periods. 288 289 With a gradual loss of protection from SARS-CoV-2 after covid-19 vaccination, many countries are now rolling out booster programmes with the aim of raising levels of immunity.
Since booster programmes began, evidence that a booster vaccine dose enhances antibody and cellular responses has accumulated. After a third dose of vaccine, neutralising antibody titres increase considerably 290–293 and, in some cases, to higher levels than after the primary two doses. 290 Additionally, boosters have also been found to increase neutralising antibody titres against the beta, gamma, delta, and omicron variants. 291 294 295 T cell response is also enhanced after a third dose. 292 296 297 Together, enhancing neutralising antibody and cellular responses with a booster vaccine dose is likely to provide a greater level of protection than relying on immunity built through a primary regimen.
The antibody and cellular responses observed after booster vaccinations have been found to correlate with increased levels of protection against SAR-CoV-2 infection and severe illness. On 30 July 2021, Israel was the first country to offer a third dose of BNT162b2 to certain groups. Subsequently, several observational studies have shown that those individuals who received a third vaccine dose were significantly less likely to be infected or have severe disease with SARS-CoV-2 than those who received two doses. 298–301 In those individuals aged 60 or older, an observational study showed that the rate of severe covid-19 and death was lower in the group that received a booster by a factor of 17.9 and 14.7, respectively, than in the group that did not receive a booster. 302 Booster doses of covid-19 vaccine have been shown to be effective against infection with the delta 303 304 and, to a lesser degree, omicron variants 75 145 146 304–306 despite the numerous mutations harboured by these variants. Overall, increasing evidence is pointing towards the benefits of booster doses of covid-19 vaccines; therefore, it is expected that booster programmes will continue to roll out across the globe. Based on current evidence, the US Centers for Disease Control and Prevention recommend that the time interval for receiving a booster after the primary regimen is five months for the BNT162b2 primary regimen, six months for the mRNA-1273 primary regimen, and two months for the Ad26.COV2.S primary regimen. 307 As the pandemic progresses and new variants emerge, variant specific vaccines could require development, with pre-clinical studies demonstrating their efficacy 308 and pharmaceutical companies, such as Pfizer, advancing in variant specific vaccine development. 146 Policy makers should also consider when vaccine boosters will be given in the future and who will receive booster doses in the long term.
Emerging treatments
As the virus becomes better understood, the therapeutic strategy against covid-19 develops. Over 2000 ongoing trials are currently assessing certain treatment strategies for covid-19. 309 Recently, antiviraldrugs including molnupiravir (Lagevrio) and nirmatrelvir/ritonavir (Paxlovid) have been approved in the UK, 310 311 US, 312 313 and Europe 314 315 for treating covid-19 in certain risk groups. Similarly, sotrovimab (Xevudy), a monoclonal antibody treatment, has recently been approved for use in treating certain patients with covid-19 in the UK, 316 US, 317 and Europe. 318 These drugs have been shown to be effective at preventing poor clinical outcomes, including death, in those individuals vulnerable to severe covid-19 infection. Other drug treatments, such as janus kinase inhibitors, corticosteroids, and anti-inflammatory drugs, have contrasting evidence to support their use; therefore, the use of specific drugs is either recommended for or against by certain treatment and management guidelines, which are discussed below.
The treatment and management of covid-19 is a continually evolving topic; however, health authorities have published and continue to update guidelines and recommendations for treating covid-19. The WHO living guideline on covid-19 and treatment is regularly updated, with the latest version (published on 14 January 2022) containing 14 recommendations on covid-19 treatment. 319 The UK National Institute for Health and Care Excellence 320 and Medicines and Healthcare products Regulatory Agency 321 provide updated guidelines on covid-19 treatment, and in Europe, the ECDC regularly publishes several guidelines providing recommendations on a range of covid-19 related topics. 322 The US National Institutes of Health 323 and Centers for Disease Control and Prevention 324 provide guidance on covid-19 treatment and management, with the Centers for Disease Control and Prevention supplying guidelines for specific groups such as employers, schools, health departments, and governments.
Considerations for the future
Novel infectious diseases and pandemics are an unpredictable but inevitable aspect of nature; therefore, we should learn from past pandemics to prepare for future ones. Firstly, the covid-19 pandemic has highlighted and amplified the existing inequalities within society, 325 with Black ethnicity, social disadvantage, and unemployment being risk factors for covid-19 infection 326 and those groups most economically deprived found to be particularly vulnerable. 327 These inequalities need resolving in order for us to be better prepared for similar situations in the future.
Next, to progress through a pandemic we should be racing against the pathogen, and not against each other. This statement becomes apparent when considering the problems faced by countries seeking out personal protective equipment, 328 and the vaccine inequity seen worldwide, 329 with developed countries often better placed to be able to purchase these items. Initiatives such as WHO’s COVAX programme are vital to protect the most vulnerable groups and reduce the global spread of disease. In October 2021, the UK government released a publication outlining where the policies implemented to reduce the impact of the covid-19 pandemic failed, and the lessons learnt from these failures. 330 The publication then presents conclusions and recommendations on how to enhance pandemic preparedness, lockdown and social distancing measures, testing and contact tracing, social care, and vaccines. In countries such as the UK, US, and much of Europe, where the covid-19 death rate has been high, steps need to be taken and lessons need to be learnt in order to be better prepared for the next pandemic. The responsibility of improving pandemic response lies with policy makers, the medical/scientific community, and the public, and will ultimately require a collaborative approach.
However, certain aspects of the response to the covid-19 pandemic have been a triumph. One major victory was the rapid development and rollout of vaccines, 331 which continue to be effective. The rollout of rapid testing and quarantine for infected individuals was also important to at least disrupt the spread of the virus, especially given that asymptomatic individuals can contribute to the spread. Furthermore, the swift identification and sharing of knowledge of SARS-CoV-2 variants between countries should be applauded. Lessons can be learnt from countries where covid-19 was controlled. In Taiwan, authorities managing the pandemic as directed by pre-covid-19 pandemic plans prompted an immediate response. Screening of all airline passengers arriving from Wuhan and high risk areas, restricting entry for non-Taiwanese citizens, 14 day quarantine periods for contacts of people with confirmed covid-19 or returning travellers, a ban on large gatherings, and widespread mask wearing were some of the quickly implemented management strategies. 332 New Zealand implemented similarly effective restrictions, with the addition of a national lockdown. 332 Many of the pandemic control components that kept infection and death numbers low in Taiwan and New Zealand could be adopted by other countries in the future and could lead to improved outcomes in terms of protecting the health of individuals and the health and wellbeing of the country. Overall, much can be learnt from the covid-19 pandemic and, as we emerge from it, the inspection of which policies failed and which succeeded is imperative.
Covid-19 remains prevalent and life threatening. Although the rollout of vaccines has been successful, attaining a high global vaccination coverage and ensuring that all healthcare systems have the capacity to cope with seasonal waves are essential. With the omicron variant highly prevalent, we must continue to learn, develop therapeutics, and remain vigilant to new variants of concern. Here, we have provided an overview of the virology of SARS-CoV-2, including the mutations harboured by variants of the virus and how these mutations effect its transmissibility and virulence. We have also discussed the vaccines that have been developed and used around the world and have provided evidence supporting the rollout of booster doses. Future priorities should focus on continuing vaccination programmes and developing variant specific vaccines as new mutations emerge. This strategy, along with the expansion of our knowledge of SARS-CoV-2 and which treatments are most successful to treat covid-19 infections will ultimately lead to favourable outcomes.
Questions for future research
How will the SARS-CoV-2 virus mutate in the future, and which mutations will give a competitive advantage that will allow the virus to inflict disease to many people?
How do we keep up with the rapidly changing SARS-CoV-2 environment and ensure that vaccines remain effective?
How do we manage the booster programme and when will future booster vaccinations be required in order to maintain high levels of immunity?
How can we learn from the current and past pandemics so that we are better prepared for the next one?
Patient involvement
The BMJ did not request patient input on this article when it was commissioned.
- covid19.trackvaccines.org
- Chan JF-W ,
- Kok K-H , et al
- Sabbatini S ,
- Bastianelli S , et al
- Pottage T ,
- Garratt I ,
- Onianwa O , et al
- Zhang L , et al
- Carraturo F ,
- Del Giudice C ,
- Morelli M , et al
- Cheung M-C ,
- Perera RAPM , et al
- McAloon C ,
- Collins Áine ,
- Hunt K , et al
- Bliddal S ,
- Banasik K ,
- Pedersen OB , et al
- Geoghegan L ,
- Arbyn M , et al
- Kronbichler A ,
- Yoon S , et al
- Hatfield KM ,
- Reddy SC , et al
- Hickey NS , et al
- Du R , et al
- Jia X , et al
- Ebinger JE ,
- Achamallah N ,
- Ji H , et al
- Williamson EJ ,
- Walker AJ ,
- Bhaskaran K , et al
- Yang F , et al
- Zhang Y , et al
- Bai T , et al
- Xiao J , et al
- Chen R , et al
- Vizcarra P ,
- Pérez-Elías MJ ,
- Quereda C , et al
- Baillie JK ,
- Wilson JF ,
- Shelton JF ,
- Shastri AJ ,
- ↵ , Ellinghaus D , Degenhardt F , et al. , Severe Covid-19 GWAS Group . Genomewide association study of severe Covid-19 with respiratory failure . N Engl J Med 2020 ; 383 : 1522-1534 . doi:10.1056/NEJMoa2020283 pmid: http://www.ncbi.nlm.nih.gov/pubmed/32558485 OpenUrl PubMed
- Hoffmann M ,
- Kleine-Weber H ,
- Schroeder S , et al
- Martin W , et al
- Prakrithi P ,
- Sundar D , et al
- Huang Q-M ,
- Zhang P-D ,
- Li Z-H , et al
- Liu Z , et al
- Masters PS ,
- Ye R , et al
- Khailany RA ,
- Buschauer R ,
- Ameismeier M , et al
- Schubert K ,
- Karousis ED ,
- Jomaa A , et al
- Lokugamage KG ,
- Rozovics JM , et al
- Snijder EJ ,
- Decroly E ,
- V'kovski P ,
- Kelly J , et al
- Lo J , et al
- Hulswit RJG ,
- de Haan CAM ,
- Uriu K , et al
- Kopecky-Bromberg SA ,
- Martínez-Sobrido L ,
- Frieman M , et al
- Xie X , et al
- Fang S , et al
- Wu D , et al
- Kreimendahl S ,
- Gordon DE ,
- Bouhaddou M , et al
- Dominguez Andres A ,
- Campos AR , et al
- Redondo N ,
- Zaldívar-López S ,
- Garrido JJ , et al
- van der Zee R ,
- de Haan CAM , et al
- Corbett KS , et al
- Tortorici MA , et al
- Azevedo M ,
- Parajuli P , et al
- Yu J , et al
- Millet JK ,
- Whittaker GR
- Hamming I ,
- Bulthuis MLC , et al
- Glowacka I ,
- Bertram S ,
- Müller MA , et al
- Yang F-Y , et al
- Duchene S ,
- Featherstone L ,
- Haritopoulou-Sinanidou M , et al
- Ecdc.europa.eu
- Imperial.ac.uk
- Luna-Muschi A ,
- Borges IC ,
- de Faria E , et al
- Ye J , et al
- Greaney AJ ,
- Hilton SK , et al
- Xu X-F , et al
- Peacock TP ,
- Goldhill DH ,
- Scudellari M
- Li J-Y , et al
- Yurkovetskiy L ,
- Pascal KE , et al
- McCarthy KR ,
- Rennick LJ ,
- Collier DA ,
- Datir R , et al
- Gamage AM ,
- Chan WOY , et al
- De Marco A ,
- Ferreira IATM , et al
- Martínez-García L ,
- Espinel MA ,
- Abreu M , et al
- Vassallo M ,
- Klotz C , et al
- McAlister FA ,
- Nabipoor M ,
- Davies NG ,
- Barnard RC , et al
- Leung GM , et al
- Cao L , et al
- Challen R ,
- Brooks-Pollock E ,
- Read JM , et al
- Paredes MI ,
- Famulare M , et al
- Werner AP ,
- Moliva JI , et al
- Gallais F ,
- Gantner P ,
- Yang G , et al
- Plante JA ,
- Liu J , et al
- Pearson CA ,
- Russell TW ,
- Wibmer CK ,
- Hermanus T , et al
- Campbell F ,
- Laurenson-Schafer H , et al
- Suleman M , et al
- Fulton BO ,
- Wloga E , et al
- de Faria E ,
- Guedes AR ,
- Oliveira MS
- Freitas ARR ,
- Beckedorff OA ,
- Cavalcanti LPdeG , et al
- Pharris A ,
- Spiteri G , et al
- Sabino EC ,
- Carvalho MPS , et al
- Dejnirattisai W ,
- Supasa P , et al
- Fischer WM ,
- Gnanakaran S , et al
- McCrone JT , et al
- Augusto G ,
- Mohsen MO ,
- Tchesnokova V ,
- Kulakesara H ,
- Larson L , et al
- Nie J , et al
- Di Giacomo S ,
- Mercatelli D ,
- Rakhimov A , et al
- Reuters.com
- Euro.who.int
- Teyssou E ,
- Delagrèverie H ,
- Visseaux B , et al
- Baidaliuk A , et al
- McMenamin J ,
- Taylor B , et al
- Fisman DN ,
- Cameroni E ,
- Woolhouse M
- Pulliam JRC ,
- van Schalkwyk C ,
- Govender N , et al
- Jackson L ,
- Khoury DS , et al
- Saunders N ,
- Maes P , et al
- Abdullahi A
- Tincho MB ,
- Imperial College Covid-19 Response Team
- ↵ BA.2 lineage report. Outbreak.info . Available: https://outbreak.info/situation-reports?pango=BA.2
- ↵ SARS-CoV-2 variants of concern and variants under investigation in England. technical briefing 35 , 2022 . Available: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1050999/Technical-Briefing-35-28January2022.pdf [Accessed 28 Jan 2022 ].
- Sacoronavirus.co.za
- Acevedo ML ,
- Alonso-Palomares L ,
- Bustamante A
- Laiton-Donato K ,
- Franco-Muñoz C ,
- Álvarez-Díaz DA , et al
- Wang R , et al
- Polack FP ,
- Thomas SJ ,
- Kitchin N , et al
- Vandebosch A , et al
- El Sahly HM ,
- Essink B , et al
- Chemaitelly H ,
- Hasan MR , et al
- Kepten E , et al
- Lopez Bernal J ,
- Andrews N ,
- Gower C , et al
- Abu-Raddad LJ ,
- Nasreen S , et al
- Nasreen S ,
- Puranik A ,
- Lenehan PJ ,
- Silvert E , et al
- Skowronski DM ,
- Setayeshgar S ,
- Zou M , et al
- Boulianne N , et al
- Charmet T ,
- Schaeffer L ,
- Grant R , et al
- Chemaitelly H , et al
- Foulkes S ,
- Saei A , et al
- Angulo FJ ,
- McLaughlin JM , et al
- Nanduri S ,
- Pilishvili T ,
- Derado G , et al
- Fowlkes A ,
- Gaglani M ,
- Groover K , et al
- Lefèvre B ,
- Tondeur L ,
- Madec Y , et al
- Pouwels KB ,
- Pritchard E ,
- Matthews PC , et al
- Williams C ,
- Al-Bargash D ,
- Macalintal C
- Fabiani M ,
- Ramigni M ,
- Gobbetto V , et al
- Moreira ED ,
- Spitzer A ,
- Henig O , et al
- Inghammar M ,
- Moghaddassi M , et al
- Cabezas C ,
- Mora-Fernandez N , et al
- Emborg H-D ,
- Valentiner-Branth P ,
- Gras-Valenti P ,
- Chico-Sanchez P ,
- Algado-Selles N
- Mason TFD ,
- Whitston M ,
- Hodgson J , et al
- Alejos B , et al
- Matthews PC ,
- Stoesser N , et al
- Regev-Yochay G ,
- Bergwerk M , et al
- Shrotri M ,
- Krutikov M ,
- Palmer T , et al
- Breeher LE ,
- Tande AJ , et al
- Thompson MG ,
- Burgess JL ,
- Naleway AL , et al
- Frenck RW ,
- June Choe Y ,
- Hwang I , et al
- Lutrick K ,
- Yoo YM , et al
- Glatman-Freedman A ,
- Bromberg M ,
- Dichtiar R , et al
- Fleming-Dutra KE ,
- Farrar JL , et al
- Martínez-Baz I ,
- Miqueleiz A ,
- Casado I , et al
- Vasileiou E ,
- Simpson CR ,
- Shi T , et al
- ecdc.europa.eu
- Rosenberg ES ,
- Dorabawila V ,
- Easton D , et al
- Newhams MM ,
- Halasa NB , et al
- Zambrano LD ,
- Olson SM , et al
- Emary KRW ,
- Golubchik T ,
- Aley PK , et al
- astrazeneca.com
- Clemens SAC ,
- Madhi SA , et al
- Baillie V ,
- Cutland CL , et al
- Govindan D ,
- Ramasubramani P
- Falsey AR ,
- Sobieszczyk ME ,
- Hirsch I , et al
- Folegatti PM ,
- Emary KRW , et al
- Costa Clemens SA ,
- Bhattacharya A ,
- Ghosh T , et al
- Murugesan M ,
- Mathews P ,
- Paul H , et al
- Alencar CH ,
- Cavalcanti LPdeG ,
- Almeida MMde , et al
- Cerqueira-Silva T ,
- Pescarini J
- Ranzani OT ,
- Leite RdosS ,
- Castilho LD
- Corchado-Garcia J ,
- Puyraimond-Zemmour D ,
- Hughes T , et al
- Barlow RS ,
- Polinski JM ,
- Weckstein AR ,
- Zemmour D ,
- Leidner D ,
- O'Brien WJ ,
- Bellino P , et al
- Yassine HM ,
- Benslimane FM , et al
- Herlihy R ,
- Bamberg W ,
- Burakoff A , et al
- Bruxvoort KJ ,
- Qian L , et al
- Wang W , et al
- Li Y , et al
- Javier S-V ,
- Percy S-B ,
- Al Kaabi N ,
- Xia S , et al
- AlHosani FI ,
- Stanciole AE ,
- AlQahtani M ,
- Bhattacharyya S ,
- Undurraga EA ,
- González C , et al
- Hitchings MDT ,
- Dorion M , et al
- Torres MSS , et al
- Paixão ES ,
- Alves FJO , et al
- Scaramuzzini Torres MS
- Tanriover MD ,
- Doğanay HL ,
- Akova M , et al
- Palacios R ,
- Batista AP ,
- Albuquerque CSN , et al
- Blackwelder W , et al
- Soneja M , et al
- Malhotra S ,
- Lodha R , et al
- Galiza EP ,
- Baxter DN , et al
- Dunkle LM ,
- Kotloff KL ,
- Gay CL , et al
- Cosgrove C , et al
- Hoosain Z , et al
- Vogler I , et al
- Arunachalam PS ,
- Scott MKD ,
- Hagan T , et al
- Falsey AR , et al
- Appelman B ,
- van der Straten K ,
- Lavell AHA , et al
- Beatty AL ,
- Peyser ND ,
- Butcher XE , et al
- Haemmerle J ,
- Velasco H , et al
- Salmerón Ríos S ,
- Mas Romero M ,
- Cortés Zamora EB , et al
- Ben-Shlomo Y , et al
- Sultan AA ,
- Ding H , et al
- Barrett JR ,
- Belij-Rammerstorfer S , et al
- Al Khames Aga QA ,
- Alkhaffaf WH ,
- Hatem TH , et al
- Le Gars M ,
- Shukarev G , et al
- Richardson SI ,
- Moyo-Gwete T , et al
- Lale A , et al
- Mukhopadhyay L ,
- Gupta N , et al
- Anderson EJ ,
- Rouphael NG ,
- Widge AT , et al
- Jackson LA ,
- Rouphael NG , et al
- Huang W , et al
- Mishra SK ,
- Pradhan SK ,
- Pati S , et al
- Tré-Hardy M ,
- Cupaiolo R ,
- Wilmet A , et al
- Navaratnam AMD ,
- Nguyen V , et al
- Mizrahi B ,
- Kalkstein N , et al
- Tre-Hardy M ,
- Almendro-Vázquez P ,
- Laguna-Goya R ,
- Ruiz-Ruigomez M , et al
- Linderman SL ,
- Moodie Z , et al
- Pan H , et al
- Wu K , et al
- Flaxman A ,
- Marchevsky NG ,
- Jenkin D , et al
- Iketani S ,
- Garcia-Beltran WF ,
- St Denis KJ ,
- Hoelzemer A , et al
- Yorsaeng R ,
- Suntronwong N ,
- Phowatthanasathian H , et al
- Munro APS ,
- Cornelius V , et al
- Madelon N ,
- Heikkilä N ,
- Bar-On YM ,
- Goldberg Y ,
- Mandel M , et al
- Cohen C , et al
- Hammerman A ,
- Sergienko R , et al
- Marudi O , et al
- Levine-Tiefenbrun M ,
- Alapi H , et al
- Hansen CH ,
- Schelde AB ,
- Moustsen-Helm IR
- Lusvarghi S ,
- Pollett SD ,
- Neerukonda SN , et al
- Koch M , et al
- Covid19-trials.com
- ema.europa.eu
- Nice.org.uk
- Blundell R ,
- Costa Dias M ,
- Chadeau-Hyam M ,
- Bodinier B ,
- Elliott J , et al
- Nielsen FBH ,
- Badiani AA , et al
- Rodgers YvanderM
- parliament.uk
- Summers J ,
- Cheng H-Y ,
- Lin H-H , et al
Supplementary materials
Supplementary data.
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
- Data supplement 1
- Data supplement 2
MY and HC contributed equally.
Contributors MY and HC performed the literature search and drafted the manuscript. HC revised and finalised the manuscript. JS reviewed and revised the manuscript. PE was responsible for the concept and design of the work. PE reviewed, revised, and finalised the manuscript. PE is the guarantor.
Funding The authors have not declared a specific grant for this research from any funding agency in the public, commercial, or not-for-profit sectors.
Competing interests We have read and understood the BMJ policy on declaration of interests and declare the following interests: PE was funded by the UK Medical Research Council and now by Higher Education Funding Council for England, received grants from Alzheimer’s Research UK, Alzheimer’s Drug Discovery Foundation, Alzheimer’s Society UK, Medical Research Council, Alzheimer’s Association US, Van-Geest Foundation, and European Union grants; PE is a consultant to Roche, Pfizer, and Novo Nordisk; received educational and research grants from GE Healthcare, Novo Nordisk, Piramal Life Science/Life Molecular Imaging, Avid Radiopharmaceuticals and Eli Lilly; and is a member of the scientific advisory board at Novo Nordisk.
Provenance and peer review Commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Read the full text or download the PDF:
You are using an outdated browser. Please upgrade your browser or activate Google Chrome Frame to improve your experience.

COVID-19 Response Case Studies
Aligned with our mission to advance the understanding and impact of microbiology, the Society reached out to our community of microbiologists to share their experiences in responding to SARS-CoV-2. We aim to showcase the perspective of scientists during the COVID-19 pandemic and the variety of roles adopted to mitigate the global crisis.
Understanding how antiviral antibodies can activate natural killer cells to improve viral control
This is the first case study in our series that comes from a group of Project Investigators (PIs). Professor Ian Humphreys, Professor Eddie Wang, Professor Richard Stanton and Professor Alan Parker are from the Viral Immunology Research Group at Cardiff University, UK. Collectively, they describe how they responded to the COVID-19 pandemic, the challenges they faced and how basic microbiology and immunology are critical components of any pandemic response.
Water-Based Epidemiology – finding a needle in a very dirty haystack
This case study comes from a research group led by David Graham, a Professor of Ecosystems Engineering at Newcastle University, UK. The account explains how the group developed an array of methods for quantifying viral load in sewage, creating the new science of water-based epidemiology, and how they employed their knowledge across various projects.
The epidemiology of COVID-19 in Ugandan settlements and neighbouring communities
This case study is written by Professor Richard Birtles, Chair of Biomedicine at the University of Salford, UK. Richard recounts his experience of establishing COVID-19 diagnostics facilities in Uganda, creating capacity for in situ SARS-CoV-2 whole genome sequencing, and how he used these new resources to clarify virus transmission routes.
Locking down labs and setting up COVID-19 testing facilities
This case study is written by Andrew Martin, a laboratory technician from the University of Salford, UK. He gives his perspective of how lockdown unfolded and how he used his time away from the lab to volunteer at the Lighthouse COVID-19 Testing labs at Alderley Park, near Manchester, UK.
Basic microbiology knowledge is crucial to the understanding of new emerging diseases
This is the second case study written by Dr Chloe James, a Lecturer in Medical Microbiology at The University of Salford, UK. In this case study, she focuses on the media engagement aspect of her response to the pandemic, including the challenges of commenting on highly politicised and emotive issues.
Searching for SARS-CoV-2 in animals
This case study was submitted by Dr Sharon Brookes, who is currently the Lead Scientist for Animal and Zoonotic Viral Diseases at Animal and Plant Health Agency (APHA). She discusses the projects that the APHA have been are involved with since the start of the pandemic, exploring SARS-CoV-2 in animals and its ability to transmit to, from and between people and animals, the development of new testing techniques and how these experiences aided the development of the APHA coronavirus team.
Using baker’s yeast to develop reference viral antigens of SAR-CoV-2
This case study is written by Professor Ed Louis of the University of Leicester, UK and Chief Scientific Officer of Phenotypeca Ltd. Ed’s account highlights the how collaboration between academia and industry can aid innovation, the challenges of establishing new working practices during lockdown and the joy of getting his hands dirty at the lab bench after many years.
Rapid development of an international radiographers online training resource
This case study is written by Dr Chloe James, a Lecturer of Medical Microbiology at the University of Salford. She recounts her experience of contributing to the University of Salford-led effort to develop new e-learning resources for radiographers caring for patients with COVID-19. She highlights the difficulties of working on this fast-paced, international effort involving over 40 people whilst working remotely.
Evaluating the utility of SARS-CoV-2 serological and rapid antigen lateral flow devices
This case study was written by Dr Suzy Pickering, a Research Fellow at King’s College London. Suzy volunteered to help at a local COVID-19 testing facility to increase testing capacity. She describes the highs and the lows of this experience, including times where she felt she was “swimming against the tide”.
From PhD student to COVID-19 testing scientist
This case study was written by Megan Taggart, a PhD candidate from Ulster University. She explains how, as a first year student, she suspended her PhD to become an RNA extraction scientist at her local COVID-19 testing labs. Megan details her time there and how it aided her development.
Saliva versus nasopharyngeal swab specimens for the detection of SARS-CoV-2
This case study comes to you from Dr Anne Wyllie, a Research Scientist at the Yale School of Public Health. She gives us a unique perspective of the pandemic from our neighbours over the water in the US. Her case study highlights the bottlenecks that were present in mass scale testing at the start of the pandemic and how she contributed to overcoming these problems.
The interface between microbiology and civil engineering
This case study is courtesy of Dr Lena Ciric, a Senior Lecturer in the Department of Civil, Environmental and Geomatic Engineering at University College London. She discusses her work on research projects during the pandemic, including a collaboration with Transport for London (TfL), and the associated challenges.
The challenge of balancing an academic career with clinical practice
This case study is written by Dr Suzy Moody, a Lecturer of Eukaryotic Microbiology at Kingston University London who’s research specialises in bioremediation of plastics. However, before she embarked on a PhD she was an Intensive Care Unit (ICU) nurse. Here, she tells us about juggling her academic responsibilities with returning to the wards to treat COVID-positive patients.
Understanding the determinants of resistance to type I interferons and IFITMs in HIV-1 envelope and SARS-CoV-2 spike
This case study is written by Helena Winstone, a PhD Candidate at King’s College London. She gives a research student’s perspective of how a project can be quickly derailed by worldwide events and can pivot to explore new avenues, as well as the frustrations involved with studying a readily-mutating virus and the associated mental health effects.
Exploring how the cell surface SARS-CoV-2 nucleocapsid protein modulates innate and adaptive immunity
This case study was written by Alberto D. López-Muñoz, a postdoctoral researcher. His account of the pandemic focuses on using immune modulation strategies to study SARS-CoV-2; volunteering for a Phase III clinical trial of the COVID-19 vaccine; the difficulties of travel restrictions and being featured in the “Postdoc Profile” series by the National Institute of Health.
The COVID-19 Genomics UK Consortium and genomic surveillance
This case study comes from Andrew Page, Head of Bioinformatics at the Quadram Institute in Norwich. He shares with us his experience of helping to establish one of the founding sequencing centres for the COVID-19 Genomics UK (COG-UK) Consortium, and how his work there aided the government and impacted on the wider society
A mid-career virologist’s pandemic!
This case study is written by Rachael Tarlinton, an Associate Professor in Veterinary Virology at the University of Nottingham. Her account of the pandemic expresses the challenges of teaching, supervising and researching throughout lockdowns; how the pressure of this affected many researchers’ mental health; and the media engagement duties of scientists.
Reverse genetics to characterise SARS-CoV-2
This case study is from Dr Maia Kavanagh Williamson, a postdoctoral research associate in Andrew Davidson’s lab at the University of Bristol. As the COVID-19 pandemic emerged, her research pivoted from RNA viruses HIV and Dengue, to using reverse genetics approaches to characterise basic properties of SARS-CoV-2. Here she details her experience of working throughout the pandemic.
The study of the airborne longevity of SARS-CoV-2
This case study is written by Henry Oswin, a PhD Candidate from Professor Jonathan Reid’s research group at the University of Bristol. He discusses how his project to develop and optimise an instrument to study the airborne longevity of E. coli rapidly adapted to study SARS-CoV-2. He details the technical challenges, as well as his own personal hurdles, that lead to significant progress in understanding the airborne longevity of the virus.
The challenges of being a virologist during the COVID-19 pandemic
This case study is from Dr Elisabetta Groppelli, a Lecturer in Global Health at St George’s University of London, who led the establishment of SARS-CoV-2 research at the Institute for Infection and Immunity. Elisabetta discusses the challenges of starting a new position and establishing a research group during a pandemic, her involvement with public engagement and the personal challenges of living through the first pandemic of our lifetime.
Stepping into public outreach during the COVID-19 pandemic
This case study was written by Dr Cheryl Walter, a Lecturer of Microbiology at the University of Hull. Cheryl discusses her involvement with public outreach, education, consultancy and the development of a SARS-CoV-2 point-of-care testing device. This work enabled her to use her virology skills and knowledge to adapt to the ongoing situation.
Screening antiviral drugs against the envelope (E) protein of SARS-CoV-2
This case study was written by Dr Gemma Swinscoe who transitioned from being a PhD student to a Postdoctoral Research Assistant in mid-2020. The study focusses on Gemma’s research investigating the envelope (E) protein from SARS-CoV-2, the challenges that she faced during the first 18 months of the pandemic and her thoughts on the influence that the pandemic has had on microbiology.
Using genomic surveillance to identify waves of SARS-CoV-2 infections in Zimbabwe
This case study was written by Professor Rob Kingsley, Group Leader at the Quadram Institute. The study focusses on Rob’s genomic surveillance work in Zimbabwe through a partnership with the National Microbiology Reference Laboratory in Harare.
Image credits:
iStock/Sakurra iStock/EmaAlmqvist iStock/Eda Hoyman iStock/PBFloyd iStock/zubada iStock/nopparit iStock/Dr_Microbe iStock/kool99 iStock/reklamlar iStock/VictoriiaIlina iStock/ChansomPantip iStock/RomoloTavani iStock/DrAfter123 iStock/fotomay iStock/metamorworks iStock/ismagilov iStock/tutti-frutti iStock/KrulUA iStock/smartboy10 iStock/Gilnature
Case report
Virology Journal welcomes well-described reports of cases that include the following:
- Unreported or unusual side effects or adverse interactions involving medications.
- Unexpected or unusual presentations of a disease.
- New associations or variations in disease processes.
- Presentations, diagnoses and/or management of new and emerging diseases.
- An unexpected association between diseases or symptoms.
- An unexpected event in the course of observing or treating a patient.
- Findings that shed new light on the possible pathogenesis of a disease or an adverse effect.
Case reports submitted to Virology Journal should make a contribution to medical knowledge and must have educational value or highlight the need for a change in clinical practice or diagnostic/prognostic approaches.
Virology Journal will not consider case reports describing preventive or therapeutic interventions, as these generally require stronger evidence.
Case reports should include relevant positive and negative findings from history, examination and investigation, and can include clinical photographs, provided these are accompanied by written consent to publish from the patient(s). Case reports should include an up-to-date review of all previous cases in the field.
Authors are encouraged to describe how the case report is rare or unusual as well as its educational and/or scientific merits in the covering letter that will accompany the submission of the manuscript.
Case report submissions will be assessed by the Editors and will be sent for peer review if considered appropriate for the journal.
Authors should seek written and signed consent to publish the information from the patients or their guardians prior to submission. The submitted manuscript must include a statement to this effect in the consent section. The editorial office may request copies of the informed consent documentation upon submission of the manuscript.
Preparing your manuscript
The information below details the section headings that you should include in your manuscript and what information should be within each section.
Please note that your manuscript must include a 'Declarations' section including all of the subheadings (please see below for more information).
Title page
The title page should:
- "A versus B in the treatment of C: a randomized controlled trial", "X is a risk factor for Y: a case control study", "What is the impact of factor X on subject Y: A systematic review, A case report etc."
- or, for non-clinical or non-research studies: a description of what the article reports
- if a collaboration group should be listed as an author, please list the Group name as an author. If you would like the names of the individual members of the Group to be searchable through their individual PubMed records, please include this information in the “Acknowledgements” section in accordance with the instructions below
- Large Language Models (LLMs), such as ChatGPT , do not currently satisfy our authorship criteria . Notably an attribution of authorship carries with it accountability for the work, which cannot be effectively applied to LLMs. Use of an LLM should be properly documented in the Methods section (and if a Methods section is not available, in a suitable alternative part) of the manuscript
- indicate the corresponding author
The Abstract should not exceed 350 words. Please minimize the use of abbreviations and do not cite references in the abstract. The abstract must include the following separate sections:
- Background: why the case should be reported and its novelty
- Case presentation: a brief description of the patient’s clinical and demographic details, the diagnosis, any interventions and the outcomes
- Conclusions: a brief summary of the clinical impact or potential implications of the case report
Keywords
Three to ten keywords representing the main content of the article.
The Background section should explain the background to the case report or study, its aims, a summary of the existing literature.
Case presentation
This section should include a description of the patient’s relevant demographic details, medical history, symptoms and signs, treatment or intervention, outcomes and any other significant details.
Discussion and Conclusions
This should discuss the relevant existing literature and should state clearly the main conclusions, including an explanation of their relevance or importance to the field.
List of abbreviations
If abbreviations are used in the text they should be defined in the text at first use, and a list of abbreviations should be provided.
Declarations
All manuscripts must contain the following sections under the heading 'Declarations':
Ethics approval and consent to participate
Consent for publication, availability of data and materials, competing interests, authors' contributions, acknowledgements.
- Authors' information (optional)
Please see below for details on the information to be included in these sections.
If any of the sections are not relevant to your manuscript, please include the heading and write 'Not applicable' for that section.
Manuscripts reporting studies involving human participants, human data or human tissue must:
- include a statement on ethics approval and consent (even where the need for approval was waived)
- include the name of the ethics committee that approved the study and the committee’s reference number if appropriate
Studies involving animals must include a statement on ethics approval and for experimental studies involving client-owned animals, authors must also include a statement on informed consent from the client or owner.
See our editorial policies for more information.
If your manuscript does not report on or involve the use of any animal or human data or tissue, please state “Not applicable” in this section.
If your manuscript contains any individual person’s data in any form (including any individual details, images or videos), consent for publication must be obtained from that person, or in the case of children, their parent or legal guardian. All presentations of case reports must have consent for publication.
You can use your institutional consent form or our consent form if you prefer. You should not send the form to us on submission, but we may request to see a copy at any stage (including after publication).
See our editorial policies for more information on consent for publication.
If your manuscript does not contain data from any individual person, please state “Not applicable” in this section.
All manuscripts must include an ‘Availability of data and materials’ statement. Data availability statements should include information on where data supporting the results reported in the article can be found including, where applicable, hyperlinks to publicly archived datasets analysed or generated during the study. By data we mean the minimal dataset that would be necessary to interpret, replicate and build upon the findings reported in the article. We recognise it is not always possible to share research data publicly, for instance when individual privacy could be compromised, and in such instances data availability should still be stated in the manuscript along with any conditions for access.
Authors are also encouraged to preserve search strings on searchRxiv https://searchrxiv.org/ , an archive to support researchers to report, store and share their searches consistently and to enable them to review and re-use existing searches. searchRxiv enables researchers to obtain a digital object identifier (DOI) for their search, allowing it to be cited.
Data availability statements can take one of the following forms (or a combination of more than one if required for multiple datasets):
- The datasets generated and/or analysed during the current study are available in the [NAME] repository, [PERSISTENT WEB LINK TO DATASETS]
- The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
- All data generated or analysed during this study are included in this published article [and its supplementary information files].
- The datasets generated and/or analysed during the current study are not publicly available due [REASON WHY DATA ARE NOT PUBLIC] but are available from the corresponding author on reasonable request.
- Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.
- The data that support the findings of this study are available from [third party name] but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of [third party name].
- Not applicable. If your manuscript does not contain any data, please state 'Not applicable' in this section.
More examples of template data availability statements, which include examples of openly available and restricted access datasets, are available here .
BioMed Central strongly encourages the citation of any publicly available data on which the conclusions of the paper rely in the manuscript. Data citations should include a persistent identifier (such as a DOI) and should ideally be included in the reference list. Citations of datasets, when they appear in the reference list, should include the minimum information recommended by DataCite and follow journal style. Dataset identifiers including DOIs should be expressed as full URLs. For example:
Hao Z, AghaKouchak A, Nakhjiri N, Farahmand A. Global integrated drought monitoring and prediction system (GIDMaPS) data sets. figshare. 2014. http://dx.doi.org/10.6084/m9.figshare.853801
With the corresponding text in the Availability of data and materials statement:
The datasets generated during and/or analysed during the current study are available in the [NAME] repository, [PERSISTENT WEB LINK TO DATASETS]. [Reference number]
If you wish to co-submit a data note describing your data to be published in BMC Research Notes , you can do so by visiting our submission portal . Data notes support open data and help authors to comply with funder policies on data sharing. Co-published data notes will be linked to the research article the data support ( example ).
All financial and non-financial competing interests must be declared in this section.
See our editorial policies for a full explanation of competing interests. If you are unsure whether you or any of your co-authors have a competing interest please contact the editorial office.
Please use the authors initials to refer to each authors' competing interests in this section.
If you do not have any competing interests, please state "The authors declare that they have no competing interests" in this section.
All sources of funding for the research reported should be declared. If the funder has a specific role in the conceptualization, design, data collection, analysis, decision to publish, or preparation of the manuscript, this should be declared.
The individual contributions of authors to the manuscript should be specified in this section. Guidance and criteria for authorship can be found in our editorial policies .
Please use initials to refer to each author's contribution in this section, for example: "FC analyzed and interpreted the patient data regarding the hematological disease and the transplant. RH performed the histological examination of the kidney, and was a major contributor in writing the manuscript. All authors read and approved the final manuscript."
Please acknowledge anyone who contributed towards the article who does not meet the criteria for authorship including anyone who provided professional writing services or materials.
Authors should obtain permission to acknowledge from all those mentioned in the Acknowledgements section.
See our editorial policies for a full explanation of acknowledgements and authorship criteria.
If you do not have anyone to acknowledge, please write "Not applicable" in this section.
Group authorship (for manuscripts involving a collaboration group): if you would like the names of the individual members of a collaboration Group to be searchable through their individual PubMed records, please ensure that the title of the collaboration Group is included on the title page and in the submission system and also include collaborating author names as the last paragraph of the “Acknowledgements” section. Please add authors in the format First Name, Middle initial(s) (optional), Last Name. You can add institution or country information for each author if you wish, but this should be consistent across all authors.
Please note that individual names may not be present in the PubMed record at the time a published article is initially included in PubMed as it takes PubMed additional time to code this information.
Authors' information
This section is optional.
You may choose to use this section to include any relevant information about the author(s) that may aid the reader's interpretation of the article, and understand the standpoint of the author(s). This may include details about the authors' qualifications, current positions they hold at institutions or societies, or any other relevant background information. Please refer to authors using their initials. Note this section should not be used to describe any competing interests.
Footnotes can be used to give additional information, which may include the citation of a reference included in the reference list. They should not consist solely of a reference citation, and they should never include the bibliographic details of a reference. They should also not contain any figures or tables.
Footnotes to the text are numbered consecutively; those to tables should be indicated by superscript lower-case letters (or asterisks for significance values and other statistical data). Footnotes to the title or the authors of the article are not given reference symbols.
Always use footnotes instead of endnotes.
Examples of the Vancouver reference style are shown below.
See our editorial policies for author guidance on good citation practice
Web links and URLs: All web links and URLs, including links to the authors' own websites, should be given a reference number and included in the reference list rather than within the text of the manuscript. They should be provided in full, including both the title of the site and the URL, as well as the date the site was accessed, in the following format: The Mouse Tumor Biology Database. http://tumor.informatics.jax.org/mtbwi/index.do . Accessed 20 May 2013. If an author or group of authors can clearly be associated with a web link, such as for weblogs, then they should be included in the reference.
Example reference style:
Article within a journal
Smith JJ. The world of science. Am J Sci. 1999;36:234-5.
Article within a journal (no page numbers)
Rohrmann S, Overvad K, Bueno-de-Mesquita HB, Jakobsen MU, Egeberg R, Tjønneland A, et al. Meat consumption and mortality - results from the European Prospective Investigation into Cancer and Nutrition. BMC Medicine. 2013;11:63.
Article within a journal by DOI
Slifka MK, Whitton JL. Clinical implications of dysregulated cytokine production. Dig J Mol Med. 2000; doi:10.1007/s801090000086.
Article within a journal supplement
Frumin AM, Nussbaum J, Esposito M. Functional asplenia: demonstration of splenic activity by bone marrow scan. Blood 1979;59 Suppl 1:26-32.
Book chapter, or an article within a book
Wyllie AH, Kerr JFR, Currie AR. Cell death: the significance of apoptosis. In: Bourne GH, Danielli JF, Jeon KW, editors. International review of cytology. London: Academic; 1980. p. 251-306.
OnlineFirst chapter in a series (without a volume designation but with a DOI)
Saito Y, Hyuga H. Rate equation approaches to amplification of enantiomeric excess and chiral symmetry breaking. Top Curr Chem. 2007. doi:10.1007/128_2006_108.
Complete book, authored
Blenkinsopp A, Paxton P. Symptoms in the pharmacy: a guide to the management of common illness. 3rd ed. Oxford: Blackwell Science; 1998.
Online document
Doe J. Title of subordinate document. In: The dictionary of substances and their effects. Royal Society of Chemistry. 1999. http://www.rsc.org/dose/title of subordinate document. Accessed 15 Jan 1999.
Online database
Healthwise Knowledgebase. US Pharmacopeia, Rockville. 1998. http://www.healthwise.org. Accessed 21 Sept 1998.
Supplementary material/private homepage
Doe J. Title of supplementary material. 2000. http://www.privatehomepage.com. Accessed 22 Feb 2000.
University site
Doe, J: Title of preprint. http://www.uni-heidelberg.de/mydata.html (1999). Accessed 25 Dec 1999.
Doe, J: Trivial HTTP, RFC2169. ftp://ftp.isi.edu/in-notes/rfc2169.txt (1999). Accessed 12 Nov 1999.
Organization site
ISSN International Centre: The ISSN register. http://www.issn.org (2006). Accessed 20 Feb 2007.
Dataset with persistent identifier
Zheng L-Y, Guo X-S, He B, Sun L-J, Peng Y, Dong S-S, et al. Genome data from sweet and grain sorghum (Sorghum bicolor). GigaScience Database. 2011. http://dx.doi.org/10.5524/100012 .
Figures, tables and additional files
See General formatting guidelines for information on how to format figures, tables and additional files.
Submit manuscript
- Editorial Board
- Manuscript editing services
- Instructions for Editors
- SNAPP Editorial Login
- Contact support for Authors
- Contact support for Editors
- Sign up for article alerts and news from this journal
- Follow us on Twitter
Annual Journal Metrics
2022 Citation Impact 4.8 - 2-year Impact Factor 3.9 - 5-year Impact Factor 1.006 - SNIP (Source Normalized Impact per Paper) 1.092 - SJR (SCImago Journal Rank)
2023 Speed 12 days submission to first editorial decision for all manuscripts (Median) 109 days submission to accept (Median)
2023 Usage 2,323,860 downloads 11,783 Altmetric mentions
- More about our metrics
Virology Journal
ISSN: 1743-422X
- Submission enquiries: [email protected]
Advertisement
SARS-CoV-2 and the nervous system: current perspectives
- Open access
- Published: 01 June 2023
- Volume 168 , article number 171 , ( 2023 )
Cite this article
You have full access to this open access article

- Amrita Pattanaik ORCID: orcid.org/0000-0002-1562-0347 1 ,
- Sushma Bhandarkar B 1 ,
- Lonika Lodha 2 &
- Srilatha Marate 1
3308 Accesses
9 Citations
50 Altmetric
Explore all metrics
SARS-CoV-2 infection frequently causes neurological impairment in both adults and children. Recent publications have described significant aspects of the viral pathophysiology associated with neurological dysfunction. In theory, neurological manifestations following SARS-CoV-2 infection may be caused directly by the effects of the virus infecting the brain or indirectly by the local and systemic immune responses against the virus. Neurological manifestations can occur during the acute phase as well as in the post-acute phase of the infection. In this review, we discuss recent literature describing the association of nervous system disorders with COVID-19.

Similar content being viewed by others
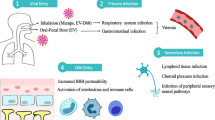
Viral meningitis: an overview

Recent Developments on Therapeutic and Diagnostic Approaches for COVID-19

Neurological Complications Following COVID-19 Vaccination
Avoid common mistakes on your manuscript.
Introduction
From the time COVID-19 was declared a pandemic by the World Health Organization (WHO), clinicians began observing neurological manifestations of both mild and severe intensity in acutely ill patients with confirmed infection [ 1 , 2 , 3 ]. Early retrospective studies from China and France revealed that a very large number of COVID-19 patients had experienced neurological complications during the period of their hospitalization [ 4 , 5 ]. However, since these reports were limited to hospitalized patients, they are not reflective of the true community-wide burden of neurological manifestations following SARS-CoV-2 infection. Furthermore, since the data used in many of these studies were extracted from the hospitals’ electronic records, there is a strong possibility that some nonspecific neurological symptoms were overlooked [ 6 ]. Considering the magnitude of the pandemic, there is a strong likelihood of neurological manifestations being experienced by a much larger population of COVID-19 patients during the course of their illness than what has been reported [ 7 ]. According to the published literature, neurological complications, such as cognitive dysfunction and encephalopathy, appear to be more debilitating than complications reported in other organ systems in COVID-19 [ 8 ]. The spectrum of neurological manifestations in SARS-CoV-2 infection likely represents multiple pathogenic pathways. Various mechanisms leading to the development of the neurological manifestations following neurotropic invasion have been proposed, including endothelial dysfunction, hyperinflammation, hypercoagulability, hypoxia, and general critical illness. There is still much to be explored in order to fully comprehend the pathogenicity of SARS-CoV-2 and its deleterious effects on the nervous system [ 7 , 9 ].
Neurological involvement has been found at different stages of SARS-CoV-2 infection – during acute infection and as post-acute sequelae manifesting in a chronic course of infection [ 9 ]. Among the many reported, the most common neurological symptoms associated with COVID-19 have been anosmia, encephalopathy, and stroke [ 10 ]. In the acute phase, infected patients frequently show nonspecific symptoms such as generalized weakness, dizziness, headache, nausea, anosmia, and dysgeusia [ 11 ]. Neurological manifestations are also frequently seen in ‘long COVID’ syndrome. Anosmia and dysgeusia, as well as neuropsychiatric symptoms, have been reported to persist for months following infection [ 12 ].
In this review, we highlight the literature focusing on clinical observations that suggest associations between SARS-CoV-2 infection and the nervous system. We also discuss the different mechanisms of neural injury that lead to various complications. Knowledge about the possible neurological manifestations of COVID-19 is vital for physicians to recognize, treat, and manage complications of the nervous system.
We searched the PubMed database for literature published between December 1, 2019, and April 1, 2023. The following search terms were included: “COVID-19”, “SARS-CoV-2”, “neuroCOVID”, “encephalopathy”, “neurological impairment”, “neurological deterioration”, “encephalitis”, “neurological manifestations”, “post-COVID symptoms”, “long COVID”, and “neurological symptoms”. Observational and interventional studies involving adult subjects were included. Commercial reports and government publications and reports were not included. Information about disease pathophysiology and clinical manifestations was extracted from the included studies.
The results are reported in a narrative form under the headings Pathogenesis: infection and neuroinflammation and Neurological manifestations. Patient-related clinical outcomes, wherever available, were also included in this summary.
Pathogenesis: infection and neuroinflammation
SARS-CoV-2, the seventh known human coronavirus, is a single-stranded enveloped RNA virus. It shares 79.5% genome sequence identity with SARS-CoV. It also shares 89–96% nucleotide sequence identity with bat coronaviruses [ 6 ]. SARS-CoV-2 binds to its receptor, angiotensin converting enzyme 2 (ACE-2), an important component of the renin-angiotensin system, to initiate replication in its host cells. The formation of the SARS-CoV-2/ACE-2 complex leads to the activation of transmembrane protease, serine 2 (TMPRSS2), which then cleaves the spike protein, allowing the SARS-CoV-2/ACE-2 complex to be internalized into the cell by endocytosis [ 13 ]. An alternate co-receptor for the virus is the membrane protein neuropilin 1 (NRP 1) [ 14 ]. After uncoating within the cell, the viral genome is used as an mRNA for translation of the viral non-structural proteins, forming a replicase-transcriptase complex (RTC) that produces subgenomic RNA for translation of the viral structural proteins. After assembly, new virions are released by exocytosis [ 15 ].
Direct invasion of the nervous system
Via olfactory nerves.
The chemosensory loss seen in COVID-19 patients in the form of anosmia, ageusia, or dysgeusia can be attributed to dysfunctional or damaged olfactory and gustatory receptors and their supporting cells or disruption of interactions with semaphorins (key molecules in olfactory and gustatory signalling pathways) [ 16 ]. There is no evidence of the expression of ACE-2 in the olfactory nerve, which seems to rule out direct neuronal damage by the virus as the cause of anosmia. However, ACE-2 receptors have been demonstrated in the olfactory mucosa by immunostaining. Olfactory epithelial sustentacular cells have been shown to express ACE-2. This has been demonstrated by single-cell sequencing and confirmed by immunostaining [ 17 , 18 , 19 ]. The spike protein (detected by immunohistochemistry) and RNA (detected by real-time PCR) of SARS-CoV-2 virus have been demonstrated in olfactory mucosa of post-mortem samples from SARS-CoV-2 infected individuals [ 20 , 21 , 22 ]. Infection of the olfactory epithelium (Fig. 1 ) can therefore account for the anosmia seen in the disease and potentially serve as a pathway of entry of the virus into the central nervous system (CNS) [ 18 , 19 ].
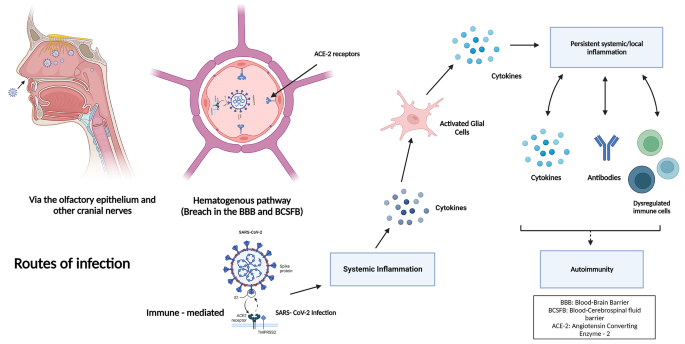
Potential routes of SARS-CoV-2 entry into the CNS (created using BioRender). A potential entry route of SARS-CoV-2 into the CNS could be via the olfactory epithelium. Another pathway of entry could be by infection of the brain capillary endothelium (hematogenous pathway). Immune responses to viral infection may result in disruption of the blood brain barrier, resulting in the creation and maintenance of an inflammatory environment in the CNS.
Via other cranial nerves
Other cranial nerves, such as the vagus, glossopharyngeal, and trigeminal nerves, may also be potential routes for the virus to enter the brain via retrograde axonal transport. These nerves get exposed to the virus during the course of infection. The vagus nerve, which is connected to the gastrointestinal tract as a part of the enteric nervous system, has an abundance of ACE-2 and NRP 1 receptors [ 23 ]. Some studies have suggested that the virus can access the CNS through peripheral fibers of the vagus nerve in the lung, similar to influenza virus [ 24 , 25 ]. A study examining brainstem neuropathology demonstrated the presence of SARS-CoV-2 in vagus nerve fibers by the use of immunohistochemistry (IHC) [ 26 ]. Another study examining the vagal and human glossopharyngeal nerves at the level of the medulla oblongata showed that ACE-2 receptors and neuropilin 1 (NRP1) co-receptors are widely expressed in axons, myelin sheaths, and nerve bundles. Together with ACE-2 and NRP1, the presence of TMPRSS2 in the supportive cells of the vagal and glossopharyngeal nerves has also been demonstrated [ 14 ]. SARS-CoV-2 dissemination has been seen in the trigeminal nerve as well, which has been implicated in anosmia and headache in patients. One hypothesis states that the virus can enter the CNS by invading the sensory axon of the trigeminal nerve in the nasal cavity. One study showed a high level of SARS-CoV-2 RNA in the trigeminal ganglion in deceased COVID-19 patients, and another post-mortem study showed axonal degeneration and cell loss in the trigeminal nerve [ 17 , 27 ]. Despite this evidence of the direct involvement of nerves in the disease process, more studies on the pathophysiology of COVID-19 need to be undertaken.
Indirect invasion of the nervous system
Hematogenous route.
A pathway that is potentially important for invasion of the CNS is infection of the brain capillary endothelium, which forms the neurovascular unit of the blood brain barrier (BBB). Studies examining post-mortem brain samples have shown the presence of virus-like particles in the capillary endothelium of the brain. The presence of SARS-CoV-2 nucleic acid in the brain has been demonstrated by polymerase chain reaction (PCR) targeting different regions of the viral genome [ 28 ]. The choroid plexus epithelial cells that form a part of the blood-cerebrospinal fluid (BCSF) barrier might also be an entry point, as evidenced in human brain organoids [ 29 ]. Evidence for the presence of the spike protein has also been found in the choroid plexus vasculature by immunostaining with anti-spike protein antibody, and by PCR. It has been seen that the infection is restricted to the lumina of the choroid plexus capillaries and medium-sized blood vessels [ 30 ].
Immune-mediated mechanisms
Immune-mediated mechanisms have been shown to play a significant role in neuroinvasion by SARS-CoV-2. Systemic inflammatory responses to infection are responsible for triggering the activation of microglial cells by excessive production of proinflammatory cytokines, including interleukins (IL-6, IL-2, IL-12, and IL-15) and tumour necrosis factor alpha (TNF-α) [ 9 ]. Some proinflammatory cytokines have been shown to have saturable mechanisms of transport from the blood to the CNS. It has been demonstrated that blood-borne proinflammatory cytokines can disrupt and traverse the blood brain barrier (BBB) to reach the cerebrospinal fluid and interstitial fluid spaces of the brain and spinal cord [ 31 ]. They, thus, play an important role in the development of neurological symptoms in patients. An increase in the levels of intrathecal interleukins (IL- 6, IL-18, IL-15) and macrophage inflammatory protein 1β (MIP-1β) has been seen in a subset of immunocompetent COVID-19 patients displaying neurological manifestations when compared to a control group of immunosuppressed SARS-CoV-2-infected patients who did not have any neurological symptoms [ 32 ]. Analysis of the CSF of a SARS-CoV-2-infected individual diagnosed with acute encephalopathy showed increased levels of pro-inflammatory cytokines, including monocyte chemoattractant protein 1 (MCP-1) [ 33 ]. Elevated levels of MCP-1 have been seen in other neuroinfectious or neuroinflammatory disorders such as neuroAIDS, bacterial meningitis, and multiple sclerosis [ 34 , 35 , 36 ].
Neuroinflammation
Neuroinflammation involves different inflammatory responses elicited against particular stimuli in the CNS. This is a result of a multitude of reactive components of the neurovascular unit (NVU) and their responses. These components include neurons, microglia, astrocytes, oligodendrocytes, and endothelial cells [ 37 ].
The various inflammatory mediators that are produced by these activated components include cytokines, chemokines, and free radicals (Fig. 2 ). These mediators contribute to the increased permeability and infiltration of immune cells across the BBB, thereby promoting neuroinflammation [ 38 , 39 , 40 ].
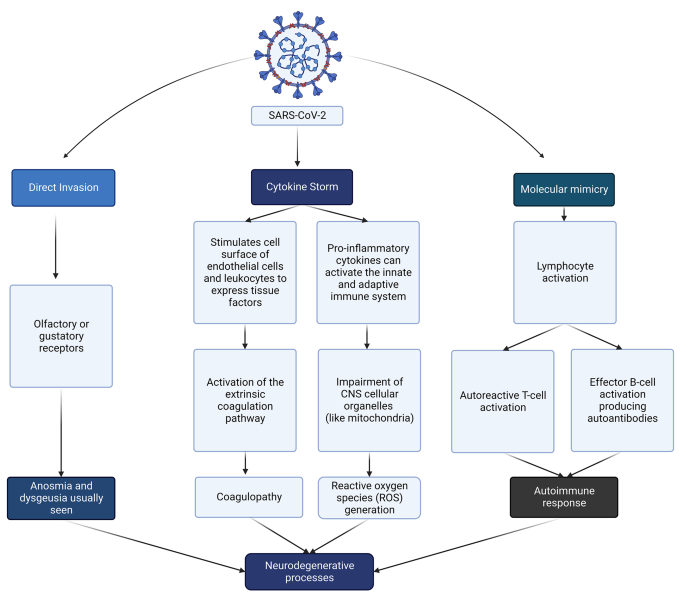
Pathogenesis of neurodegenerative processes in COVID-19 (created using BioRender). Neurodegenerative processes can be observed as a result of direct invasion via the olfactory or gustatory receptors, coagulopathy, generation of reactive oxygen species (ROS), or induction of autoimmunity.
It has been found that SARS-CoV-2-infected patients with acute neurological complications have elevated levels of proinflammatory cytokines such as IL-6, IL-18, and IL-8 when compared to healthy controls [ 41 ]. Similarly, another study reported increased levels of proinflammatory cytokines (IL-6, IL-8, and TNF-α) in CSF from a SARS-CoV-2-infected individual with akinetic mutism. It was also shown that cytokine levels decreased with the recovery of the individual [ 42 ]. An in vitro study in which human brain microvascular endothelial cells (BMVECs) were exposed to SARS-CoV-2 spike protein showed that there was an association between a reduction in the expression of tight-junction and an increase in the levels of the cytokines IL-6, IL-10, and TNF-α [ 43 ].
Other studies have shown that, in anosmia, the loss of olfactory function could be correlated with infection of the olfactory epithelium and increased expression of cytokines such as IL-6 [ 10 , 44 ]. The regeneration of nasal epithelial cells may be compromised, as the olfactory mucosa is sensitive to cytokines [ 45 ]. Proinflammatory cytokines such as IL-6 can potentially activate Toll-like receptors, which may lead to inflammation of taste buds and, thereby, a loss of taste [ 46 , 47 ].
Oxidative stress, a significant inflammatory response that has been implicated in neuroAIDS in human immunodeficiency virus (HIV) infection, is suggested to play a role in the pathogenesis of neurological manifestations of COVID-19. An in vitro study performed on human microglia treated with the SARS-CoV-2 spike protein showed that there was an increase in mitochondrial respiration, leading to the production of ROS, and increased oxidative stress arising as a result of an imbalance between ROS production and the body’s ability to detoxify the reactive intermediates. This might contribute to the neurodegenerative process [ 48 , 49 , 50 ]. Neurodegenerative disease may also occur due to the activation of autoimmune responses by molecular mimicry of self-antigens by viral antigens. An example of this is Guillain-Barre Syndrome (GBS), an autoimmune disease that has been seen in some COVID-19 patients [ 51 , 52 ].
Coagulopathy has been observed in patients with severe COVID-19. The onset of stroke in COVID-19 could be due to hypercoagulability and vasculitis. In acute infections, mediators of inflammation such as tissue factors may induce hypercoagulation [ 46 ]. Tissue factors can act as receptors for factor VII. The expression of tissue factor on the cell surface of endothelium and leukocytes can be stimulated by proinflammatory cytokines such as TNF-α by the extrinsic coagulation pathway. A prothrombotic state and vasculitis could be attributed to elevation of levels of adhesion molecules, cytokines, angiotensin II, and D-dimer and a decrease in fibrinolysis, which has been associated with the disseminated intravascular coagulation (DIC) seen in severe COVID-19 [ 47 , 53 ]. Additionally, the proinflammatory cytokine IL-6 has been shown to be responsible for stimulation of platelet production, tissue factor gene expression, and fibrinogen production by endothelial and monocyte cell types [ 54 ]. Furthermore, damage to endothelial cells by cytokine storm can lead to the production of phosphatidylserine (PS), which can promote thrombin production. Release of plasminogen activator inhibitor type 1 (PAI-1) by damaged endothelial cells can inhibit the fibrinolytic system, leading to thrombosis [ 55 ]. Systemic coagulopathy and vasculopathy result in neurological manifestations such as encephalopathy and delirium seen in a SARS-CoV-2 infection [ 10 ].
The activation of glial cells (astrocytes, oligodendrocytes, and microglial cells) due to systemic infection modulates neuroinflammatory responses [ 37 ]. Microglial cells, when activated, polarise to the M1 phenotype. This phenotype mediates a proinflammatory response which involves an increase in proinflammatory mediators such as TNF-α, IL-β, IL-6, and reactive oxygen species (ROS) [ 37 , 56 ]. This proinflammatory state can lead to the activation of astrocytes [ 37 ] or lead to the destruction of microglia [ 56 ]. A post-mortem study done on the brainstems and olfactory bulbs of individuals who had succumbed to SARS-CoV-2 infection revealed high microglial immune activation with microglial nodules and immune cell clusters (such as CD8 + T cells) associated with axonal damage [ 57 ]. Activated microglia can produce IL-1 and TNF-α, which can activate astrocytes, which then produce inflammatory factors such as TNF-α, nitric oxide (NO), and ROS. This mutual interaction between microglia and astrocytes amplifies neuroinflammation [ 37 , 58 ]. Another stress factor that can contribute to disruption of the integrity of the BBB is hypoxia, which can lead to the infiltration of immune cells and proinflammatory cytokines into the brain [ 51 ]. Neurological manifestations such as stroke and meningoencephalitis may be due the cytokine storm (Fig. 2 ) induced by SARS-CoV-2 infection [ 46 ].
Neurological manifestations of SARS-CoV-2
Parainfectious/acute neurological manifestations.
Various acute neurological manifestations have been associated with COVID-19 (Table 1 ). The most common nonspecific symptoms of the nervous system reported in COVID-19 patients include olfactory and gustatory dysfunction presenting as anosmia, dysgeusia, headache, and fatigue. A meta-analysis of 350 studies with 145,721 subjects found that the pooled prevalence of taste and smell dysfunction were 21% and 19%, respectively [ 59 ]. In most cases, these symptoms were the initial manifestations of the illness and were not associated with nasal discharge or congestion. However, taste and smell disturbances were rarely the only COVID-19 symptoms and were accompanied by other manifestations. Anosmia has been reported more commonly in younger individuals than in older ones. Also, it has been seen more frequently in women than in men [ 60 ]. Smell and taste disorders resolve spontaneously without the requirement for any specific management in most patients with COVID-19. However, 10–20% of these patients have serious or long-term deficits [ 59 , 61 ].
Headache, with an incidence of 25–47%, has been reported as an acute-phase manifestation of COVID-19. Fatigue was another common, debilitating symptom during acute illness. The reported prevalence was around 27–32%. However, it is likely to have been underdiagnosed due to the subjectivity of reporting [ 71 , 72 , 73 ].
Other reported cranial nerve dysfunctions in COVID-19 patients include oculomotor dysfunction, hearing loss, facial palsy, ocular neuropathies, and lower cranial-nerve abnormalities. During or after infection, sudden sensorineural hearing loss (unilateral or bilateral) with an incidence of 13% and persistent tinnitus with an incidence of 15% have been reported relatively frequently [ 71 , 74 ].
Another nonspecific neurological manifestation commonly seen in COVID-19 patients is myalgia, which has been documented and reported in 22–63% of patients. It has been reported in mild as well as severe COVID-19 [ 61 ]. Increased levels of creatinine kinase (CK) have been seen in more-severe cases. Myopathy has also been reported in some patients during the acute phase of infection [ 60 ]. It is not clear if this is due to the direct effect of the virus on myocytes or due to the local and/or systemic immune response against the invading virus [ 75 ].
Among the more-specific neurological symptoms, encephalopathy is commonly diagnosed in patients with COVID-19. Different studies have shown its prevalence to be 8%-12% [ 62 , 76 ]. The term “encephalopathy” includes altered consciousness, delirium, agitation, confusion, or coma. Signs of encephalopathy are seen in most critically ill COVID-19 patients. Some of the risk factors associated with encephalopathy are older age, smoking, prior history of neurological derangement, diabetes, chronic kidney disease, cerebral vasculitis, dyslipidaemia, cardiac failure, and hypertension. Of the patients admitted to an intensive care unit (ICU), 60% present with agitation and delirium. In patients older than 60 years, acute confusion or delirium was seen with a pooled prevalence of 34% and was associated with higher mortality [ 9 , 59 ]. Occasional cases of encephalitis and meningitis have also been reported in COVID-19 patients. The incidence of encephalitis in COVID-19 is less than 1% but can be as high as 6–7% in severe disease [ 77 ]. Patients with encephalitis or meningitis due to suspected SARS-CoV-2 infection have presented with a wide range of typical presentations (signs of meningeal irritation, altered sensorium) and atypical presentations (like seizure, akinetic mutism, psychosis, oculocephalic reflex, catatonia, coma) [ 42 , 78 , 79 , 80 , 81 , 82 ].
New-onset seizures are one of the important acute-phase manifestations of neurological dysfunction reported during SARS-CoV-2 infection. A recent study concluded that acute seizures occurred in less than 5% of the hospitalized COVID-19 patients [ 72 ]. Acute stroke or encephalitis are frequently associated with seizures. Various studies have concluded that most of these seizures developed in the absence of a prior diagnosis of epilepsy [ 83 ].
An association between COVID-19 and cerebrovascular disease has been convincingly demonstrated, with manifestations including ischemic stroke, intracerebral thrombosis, and intracerebral haemorrhage. These manifestations have been observed not only in older patients with multiple significant cerebrovascular risk factors but also in young patients without any comorbidities. A meta-analysis involving 18,258 COVID-19 patients showed that the pooled prevalence of cerebral ischemia was 2.9% and that of cerebral thrombosis was 2.2% [ 62 ]. Morassi et al., in a case series, reported biochemical evidence of coagulopathy in more than 65% of patients with COVID-19 [ 84 ]. Stroke usually developed within a month of onset of the symptoms of COVID-19. In different studies, it was seen that SARS-CoV-2 infection was an independent risk factor for stroke in hospitalized patients [ 85 , 86 , 87 , 88 ].
Psychiatric disorders have been one of the significant CNS disturbances described during the pandemic. A study from the United States retrospectively reported psychiatric manifestations in COVID-19 patients within the first 3 months of infection. These patients did not have any previous history of psychiatric disorders. The most frequent disorders reported in that study were insomnia, anxiety, and dementia. Ten to 38% of cases of depression and/or anxiety associated with SARS-CoV-2 infection occurred during the acute phase of illness. In 5–13% of the cases, symptoms persisted even after the resolution of the infection [ 71 , 72 , 73 , 89 ].
Post-acute neurological manifestations
A few months into the SARS-CoV-2 pandemic, anecdotal reports from survivors of the illness started to emerge from social media and patient support groups, complaining of non-resolution of symptoms or protracted course of illness for weeks to months [ 90 ]. In the initial reports, the constellation of persistent symptoms in COVID-19 survivors was labelled as “long-haul COVID” or “long-tail COVID” by the mainstream media. It was reported that after the resolution of acute respiratory and febrile illness, patients suffered from a wide spectrum of systemic and organ-system-specific symptoms that persisted long after microbiological recovery (i.e., a negative PCR test) [ 91 ].
However, research data were scarce at that time, and there was no concrete definition or diagnostic criteria for this syndrome. Subsequently, in October 2021, the WHO formally recognised this post-COVID-19 condition by formulating a clinical case definition. The definition states that
Post COVID-19 condition occurs in individuals with a history of probable or confirmed SARS-CoV-2 infection, usually 3 months from the onset of COVID-19 with symptoms that last for at least 2 months and cannot be explained by an alternative diagnosis. Common symptoms include fatigue, shortness of breath, cognitive dysfunction but also others and generally have an impact on everyday functioning. Symptoms may be new onset following initial recovery from an acute COVID-19 episode or persist from the initial illness. Symptoms may also fluctuate or relapse over time .
Additionally, a code was assigned in the tenth revision of the International Classification of Diseases (ICD-10) for this condition [ 92 ].
Currently, several different terms are being used for post-COVID-19 conditions, such as post-acute sequelae of SARS-CoV-2 infection (PASC), post-acute COVID-19 syndrome (PACS), long COVID, persisting COVID, and post-COVID syndrome, among others [ 12 ]. The post-acute manifestations of COVID-19 are quite diverse, including, but not limited to, systemic, neurological, cardiovascular, respiratory, gastrointestinal, renal, immunological, and musculoskeletal dysfunctions [ 93 ].
Prevalence and duration of long COVID
The overall global prevalence of PACS has been reported to be 0.37 at 30 days, 0.25 at 60 days, 0.32 at 90 days, and 0.49 at 120 days postinfection, according to a recent meta-analysis [ 94 ]. Higher prevalence rates of 63.2% at 30 days, 71.9% at 60 days, and 45.9% at ≥ 90 days after onset of illness have also been reported [ 95 ].
In a large electronic health record review of more than 200,000 patients diagnosed with COVID-19, it was found that 33.62% of these patients presented with neurological or psychiatric sequelae within 6 months of acute infection [ 96 ]. It is important to note that there may be substantial overlap of acute and post-acute neurological manifestations of COVID-19. Therefore it is important to exclude acute infection in order to identify post-infectious sequelae [ 12 ].
The average duration from acute COVID-19 infection to post-infectious neurological sequelae has been found in a meta-analysis of 55 such cases to be 33.2 days. The conditions reported to occur, in descending order of frequency, were Guillain-Barre Syndrome (GBS), stroke, optic neuritis, and encephalitis. Less frequently, transverse myelitis, neuromyopathy/neuropathy, encephalopathy, Parkinsonism, status epilepticus, Bell’s palsy, vestibulocochlear neuritis, opsoclonus myoclonus syndrome, and myopathy were also reported to occur [ 97 ].
Studies have been carried out to evaluate the duration of persistence of long COVID symptoms, and it has been demonstrated that 37.8% of the patients studied experienced symptoms until the end of one year following acute infection [ 98 ]. However, the proportion of survivors with sequelae has been shown to decrease over time, from 68% at 6 months postinfection to 55% at two years postinfection. At the end of two years, the health status of these patients was seen to deteriorate considerably compared to the general population [ 99 ]. This long-persisting illness, i.e., at 12–18 months postinfection, has been dubbed “very long COVID” in a study with a reported prevalence of 61% in the 121 patients studied [ 100 ].
Several investigators have compared the incidence of persistent manifestations in COVID-19 survivors with other control groups, leading to disparate conclusions. A significantly higher prevalence of persistent symptoms or worse health outcomes has been demonstrated in COVID-19 patients when compared to uninfected controls, influenza-virus-infected controls, and controls with other respiratory tract infections [ 96 , 101 , 102 ]. In contrast, a few investigators have found no evidence of varying recovery rates in olfactory dysfunction in COVID-19 PCR-positive and PCR-negative patients [ 103 ]. Also, no significant difference in the prevalence of neurological and cognitive deficits has been seen in COVID-19 cases and uninfected controls [ 104 ].
Risk factors for long COVID
Several risk factors associated with the occurrence of post-infectious sequelae have been identified. As is evident from several studies, long COVID has been seen more frequently in females than in males [ 105 , 106 , 107 , 108 , 109 , 110 , 111 , 112 , 113 , 114 , 115 , 116 ]. An important predictor of the development of long COVID in survivors is the older age of the patient [ 105 , 108 , 111 , 116 ]. However, there are contradictory reports of the rate of long COVID being slightly higher in young adults [ 110 , 115 ]. Several pre-existing conditions that are associated with long COVID have been identified, such as obesity, chronic pulmonary disease, alcohol or tobacco consumption, constitutional neuropsychiatric symptoms, and others, such as hypertension, diabetes, asthma or chronic obstructive pulmonary disease (COPD), immunological disorders, hematological disorders, and malignancies [ 105 , 106 , 108 , 109 , 112 , 114 , 115 , 116 ].
The course of acute COVID-19 infection also affects the probability of developing long COVID. In particular, a severe acute illness, longer hospitalization, and ICU stay are important predictors of the occurrence of long COVID [ 105 , 107 , 109 , 110 , 111 , 113 , 114 , 116 , 117 ]. The occurrence of neurological complications, such as myalgia, tachycardia, dyspnoea, chest pain, congestion, and depression during acute infection is a predictor of long COVID [ 105 , 107 , 112 , 117 ]. Administration of corticosteroids, antibiotics, or intravenous immunoglobulins has also been found to be associated with increased incidence of long COVID [ 105 , 112 , 116 , 117 ].
Which variant of SARS-CoV-2 caused the initial acute infection does not appear to affect the development of long COVID, as demonstrated in a study involving 57,727 SARS-CoV-2-positive individuals in which the incidence of long COVID was compared between patients infected with the Delta and Omicron variants [ 118 ].
Neurological manifestations of long COVID
Generalized fatigue, weakness, or malaise have consistently been found to be the most common sequelae of SARS-CoV-2 infection [ 71 , 94 , 119 ]. Additionally, a wide array of chronic neurological complications have been reported, involving the central nervous system (for example, headache, fatigue, confusion/‘brain fog’, insomnia, or cognitive impairment, and neuropsychiatric manifestations such as depression and anxiety, dizziness, or dysautonomia) as well as the peripheral nervous system (for example, sensorimotor deficits, myopathies, muscle weakness, myalgias, disturbances in taste and/or smell, or sensorineural hearing loss/tinnitus) [ 12 ]. The wide spectrum of reported neurological and psychiatric symptoms and their variable prevalence rates seen in PACS are shown in Online Resource 1.
The progression of post-COVID sequelae over time was described in a meta-analysis of 63 studies, which reported that, from 3 to 6 months postinfection, fatigue (32%), dyspnoea (25%), sleep disorder (24%), and difficulty in concentrating (22%) were the most prevalent symptoms, while effort intolerance (45%), fatigue (36%), sleep disorder (29%), and dyspnoea (25%) were the predominant symptoms observed between 6 and 9 months postinfection. Similarly, between 9 and 12 months postinfection, fatigue (37%) and dyspnoea (21%) were the predominant symptoms. Fatigue continued to persist in 41% of the patients studied beyond 12 months [ 120 ]. Consistent with these findings, a cohort study of 121 hospitalized COVID-19 patients showed that fatigue was the most commonly reported symptom (reported by 50% of the patients), followed by dyspnoea (42%) and memory dysfunction (34%). Several other symptoms were also observed, including confusion, paresthesia, anxiety, depression, disturbed sleep, and muscle pain [ 100 ].
The high prevalence rates of neurological sequelae of COVID-19 present a major public health challenge. There is a pressing need to standardize the definition and diagnostic criteria for PACS and conduct large-scale prospective studies in order to better understand these symptoms and their risk factors. This will also help physicians to better care for these patients and possibly prevent the occurrence of further complications [ 71 ].
Conclusions
This review highlights neurological involvement in SARS-CoV-2 infection and the many neurological presentations seen during and after the course of the disease. A wide spectrum of neurological syndromes has been reported in COVID-19 patients. The probable pathogenesis of the COVID-19-associated neurological disorders is multifaceted. Across the wide array of these neurological sequelae, there lies a common element of neuroinvasion and systemic inflammation. A few of the well-studied pathogenetic mechanisms include hypoxia, overproduction of cytokines, microvascular pathology, and glial cell dysfunction. Acute neurological manifestations are mostly nonspecific. Post-acute neurological manifestations are diverse, not well defined, and may occur individually or together with other multiorgan dysfunctions. The data pertaining to long COVID are constantly evolving and need to be screened more thoroughly for a better understanding of the disease process and outcome.
It is expected that this review will help healthcare professionals to be aware of the wide constellation of neurological manifestations following SARS-CoV-2 infection for appropriate diagnosis and management and that this will reduce the morbidity in this cohort of patients. Further studies are needed for establishing a definitive association of such symptoms with COVID-19 and also for a better comprehension of the underlying pathophysiological mechanisms.
Data availability
The data used for this review are publicly available from the sources described in the methods section.
Huang C, Huang L, Wang Y et al (2021) 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. The Lancet 397:220–232. https://doi.org/10.1016/S0140-6736(20)32656-8
Article CAS Google Scholar
Majolo F, da Silva GL, Vieira L et al (2021) Neuropsychiatric Disorders and COVID-19: What We Know So Far. https://doi.org/10.3390/ph14090933 . Pharmaceuticals 14:
Marshall M (2020) How COVID-19 can damage the brain. Nature 585:342–343. https://doi.org/10.1038/d41586-020-02599-5
Article CAS PubMed Google Scholar
Mao L, Jin H, Wang M et al (2020) Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol 77:683–690. https://doi.org/10.1001/jamaneurol.2020.1127
Article PubMed Google Scholar
Helms J, Kremer S, Merdji H et al (2020) Neurologic Features in Severe SARS-CoV-2 Infection. N Engl J Med 382:2268–2270. https://doi.org/10.1056/NEJMc2008597
Pezzini A, Padovani A (2020) Lifting the mask on neurological manifestations of COVID-19. Nat Reviews Neurol 16:636–644. https://doi.org/10.1038/s41582-020-0398-3
Samuel J, Ahmad DJA, Chaim M, Feigen JP, Vazquez, Andrew J, Kobets (2022) Neurological Sequelae of COVID-19. JIN 21:77-null. https://doi.org/10.31083/j.jin2103077
Graham EL, Clark JR, Orban ZS et al (2021) Persistent neurologic symptoms and cognitive dysfunction in non-hospitalized Covid-19 “long haulers. Ann Clin Transl Neurol 8:1073–1085. https://doi.org/10.1002/acn3.51350
Article CAS PubMed PubMed Central Google Scholar
Brola W, Wilski M (2022) Neurological consequences of COVID-19. Pharmacol Rep 74:1208–1222. https://doi.org/10.1007/s43440-022-00424-6
Article PubMed PubMed Central Google Scholar
Solomon T (2021) Neurological infection with SARS-CoV-2 — the story so far. Nat Reviews Neurol 17:65–66. https://doi.org/10.1038/s41582-020-00453-w
Aschman T, Mothes R, Heppner FL, Radbruch H (2022) What SARS-CoV-2 does to our brains. Immunity 55:1159–1172. https://doi.org/10.1016/j.immuni.2022.06.013
Stefanou M-I, Palaiodimou L, Bakola E et al (2022) Neurological manifestations of long-COVID syndrome: a narrative review. Therapeutic Adv Chronic Disease 13:20406223221076890. https://doi.org/10.1177/20406223221076890
Article Google Scholar
Hoffmann M, Kleine-Weber H, Schroeder S et al (2020) SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181:271–280e8. https://doi.org/10.1016/j.cell.2020.02.052
Vitale-Cross L, Szalayova I, Scoggins A et al (2022) SARS-CoV-2 entry sites are present in all structural elements of the human glossopharyngeal and vagal nerves: Clinical implications. https://doi.org/10.1016/j.ebiom.2022.103981 . eBioMedicine 78:
Bratosiewicz-Wąsik J (2022) Neuro-COVID-19: an insidious virus in action. Neurol Neurochir Pol 56:48–60. https://doi.org/10.5603/PJNNS.a2021.0072
Xu W, Sunavala-Dossabhoy G, Spielman AI (2022) Chemosensory loss in COVID‐19. Oral Dis. https://doi.org/10.1111/odi.14300
Meinhardt J, Radke J, Dittmayer C et al (2021) Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat Neurosci 24:168–175. https://doi.org/10.1038/s41593-020-00758-5
Chen M, Shen W, Rowan NR et al (2020) Elevated ACE2 expression in the olfactory neuroepithelium: implications for anosmia and upper respiratory SARS-CoV-2 entry and replication. bioRxiv: the preprint server for biology 2020.05.08.084996
Brann DH, Tsukahara T, Weinreb C et al (2020) Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci Adv 6:eabc5801. https://doi.org/10.1126/sciadv.abc5801
Fabbri VP, Foschini MP, Lazzarotto T et al (2020) Brain ischemic injury in COVID-19‐infected patients: a series of 10 post‐mortem cases. Brain Pathol 31:205–210. https://doi.org/10.1111/bpa.12901
Puelles VG, Lütgehetmann M, Lindenmeyer MT et al (2020) Multiorgan and Renal Tropism of SARS-CoV-2. N Engl J Med 383:590–592. https://doi.org/10.1056/NEJMc2011400
Al-Dalahmah O, Thakur KT, Nordvig AS et al (2020) Neuronophagia and microglial nodules in a SARS-CoV-2 patient with cerebellar hemorrhage. Acta Neuropathol Commun 8:147. https://doi.org/10.1186/s40478-020-01024-2
Burks SM, Rosas-Hernandez H, Alejandro Ramirez-Lee M, Brain et al (2021) Behav Immun 95:7–14. https://doi.org/10.1016/j.bbi.2020.12.031
Lima M, Siokas V, Aloizou A-M et al (2020) Unraveling the Possible Routes of SARS-COV-2 Invasion into the Central Nervous System. Curr Treat Options Neurol 22:37. https://doi.org/10.1007/s11940-020-00647-z
Matsuda K, Park CH, Sunden Y et al (2004) The Vagus Nerve is One Route of Transneural Invasion for Intranasally Inoculated Influenza A Virus in Mice. Vet Pathol 41:101–107. https://doi.org/10.1354/vp.41-2-101
Bulfamante G, Bocci T, Falleni M et al (2021) Brainstem neuropathology in two cases of COVID-19: SARS-CoV-2 trafficking between brain and lung. J Neurol 268:4486–4491. https://doi.org/10.1007/s00415-021-10604-8
von Weyhern CH, Kaufmann I, Neff F, Kremer M (2020) Early evidence of pronounced brain involvement in fatal COVID-19 outcomes. The Lancet 395:e109. https://doi.org/10.1016/S0140-6736(20)31282-4
Paniz-Mondolfi A, Bryce C, Grimes Z et al (2020) Central nervous system involvement by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). J Med Virol 92:699–702. https://doi.org/10.1002/jmv.25915
Zhang L, Zhou L, Bao L et al (2021) SARS-CoV-2 crosses the blood–brain barrier accompanied with basement membrane disruption without tight junctions alteration. Signal Transduct Target Therapy 6:337. https://doi.org/10.1038/s41392-021-00719-9
Gomes I, Karmirian K, Oliveira JT et al (2021) SARS-CoV-2 infection of the central nervous system in a 14-month-old child: A case report of a complete autopsy. Lancet Reg Health - Americas 2:100046. https://doi.org/10.1016/j.lana.2021.100046
Banks WA, Kastin AJ, Broadwell RD (1995) Passage of Cytokines across the Blood-Brain Barrier. Neuroimmunomodulation 2:241–248. https://doi.org/10.1159/000097202
Normandin E, Holroyd KB, Collens SI et al (2021) Intrathecal inflammatory responses in the absence of SARS-CoV-2 nucleic acid in the CSF of COVID-19 hospitalized patients. J Neurol Sci 430:120023. https://doi.org/10.1016/j.jns.2021.120023
Farhadian S, Glick LR, Vogels CBF et al (2020) Acute encephalopathy with elevated CSF inflammatory markers as the initial presentation of COVID-19. BMC Neurol 20:248. https://doi.org/10.1186/s12883-020-01812-2
Sozzani S, Introna M, Bernasconi S et al (1997) MCP-1 and CCR2 in HIV infection: regulation of agonist and receptor expression. J Leukoc Biol 62:30–33. https://doi.org/10.1002/jlb.62.1.30
Mastroianni CM, Lancella L, Mengoni F et al (2001) Chemokine profiles in the cerebrospinal fluid (CSF) during the course of pyogenic and tuberculous meningitis. Clin Exp Immunol 114:210–214. https://doi.org/10.1046/j.1365-2249.1998.00698.x
Mahad DJ, Ransohoff RM (2003) The role of MCP-1 (CCL2) and CCR2 in multiple sclerosis and experimental autoimmune encephalomyelitis (EAE). Semin Immunol 15:23–32. https://doi.org/10.1016/S1044-5323(02)00125-2
Liu L, Liu J, Bao J et al (2020) Interaction of Microglia and Astrocytes in the Neurovascular Unit. Front Immunol 11. https://doi.org/10.3389/fimmu.2020.01024
DiSabato DJ, Quan N, Godbout JP (2016) Neuroinflammation: the devil is in the details. J Neurochem 139:136–153. https://doi.org/10.1111/jnc.13607
Yang Q, Zhou J (2019) Neuroinflammation in the central nervous system: Symphony of glial cells. Glia 67:1017–1035. https://doi.org/10.1002/glia.23571
Sivandzade F, Alqahtani F, Cucullo L (2020) Traumatic Brain Injury and Blood–Brain Barrier (BBB): Underlying Pathophysiological Mechanisms and the Influence of Cigarette Smoking as a Premorbid Condition. Int J Mol Sci 21. https://doi.org/10.3390/ijms21082721
Guasp M, Muñoz-Sánchez G, Martínez-Hernández E et al (2022) CSF Biomarkers in COVID-19 Associated Encephalopathy and Encephalitis Predict Long-Term Outcome. Front Immunol 13. https://doi.org/10.3389/fimmu.2022.866153
Pilotto A, Odolini S, Masciocchi S et al (2020) Steroid-Responsive Encephalitis in Coronavirus Disease 2019. Ann Neurol 88:423–427. https://doi.org/10.1002/ana.25783
Reynolds JessicaL, Mahajan SD (2021) SARS-COV2 Alters Blood Brain Barrier Integrity Contributing to Neuro-Inflammation. J Neuroimmune Pharmacol 16:4–6. https://doi.org/10.1007/s11481-020-09975-y
de Melo GD, Lazarini F, Levallois S et al (2021) COVID-19-related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci Transl Med 13:eabf8396. https://doi.org/10.1126/scitranslmed.abf8396
Cooper KW, Brann DH, Farruggia MC et al (2020) COVID-19 and the Chemical Senses: Supporting Players Take Center Stage. Neuron 107:219–233. https://doi.org/10.1016/j.neuron.2020.06.032
Galea M, Agius M, Vassallo N (2022) Neurological manifestations and pathogenic mechanisms of COVID-19. Neurol Res 44:571–582. https://doi.org/10.1080/01616412.2021.2024732
Allegra A, Innao V, Allegra AG, Musolino C (2020) Coagulopathy and thromboembolic events in patients withSARS-CoV-2 infection: pathogenesis and management strategies. Ann Hematol 99:1953–1965. https://doi.org/10.1007/s00277-020-04182-4
Clough E, Inigo J, Chandra D et al (2021) Mitochondrial Dynamics in SARS-COV2 Spike Protein Treated Human Microglia: Implications for Neuro-COVID. J Neuroimmune Pharmacol 16:770–784. https://doi.org/10.1007/s11481-021-10015-6
Chen X, Guo C, Kong J (2012) Oxidative stress in neurodegenerative diseases. Neural Regen Res 7:376–385. https://doi.org/10.3969/j.issn.1673-5374.2012.05.009
Mollace V, Nottet HSLM, Clayette P et al (2001) Oxidative stress and neuroAIDS: triggers, modulators and novel antioxidants. Trends Neurosci 24:411
Mahboubi Mehrabani M, Karvandi MS, Maafi P, Doroudian M (2022) Neurological complications associated with Covid-19; molecular mechanisms and therapeutic approaches. Rev Med Virol 32:e2334. https://doi.org/10.1002/rmv.2334
Sriwastava S, Kataria S, Tandon M et al (2021) Guillain Barré Syndrome and its variants as a manifestation of COVID-19: A systematic review of case reports and case series. J Neurol Sci 420:117263. https://doi.org/10.1016/j.jns.2020.117263
Tang N, Li D, Wang X, Sun Z (2020) Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost 18:844–847. https://doi.org/10.1111/jth.14768
Kerr R, Stirling D, Ludlam CA (2001) Interleukin 6 and Haemostasis. Br J Haematol 115:3–12. https://doi.org/10.1046/j.1365-2141.2001.03061.x
Zhang S, Zhang J, Wang C et al (2021) COVID–19 and ischemic stroke: Mechanisms of hypercoagulability (Review). Int J Mol Med 47:21. https://doi.org/10.3892/ijmm.2021.4854
Jeong GU, Lyu J, Kim K-D et al (2022) SARS-CoV-2 Infection of Microglia Elicits Proinflammatory Activation and Apoptotic Cell Death. Microbiol Spectr 10:e01091–e01022. https://doi.org/10.1128/spectrum.01091-22
Schwabenland M, Salié H, Tanevski J et al (2021) Deep spatial profiling of human COVID-19 brains reveals neuroinflammation with distinct microanatomical microglia-T-cell interactions. Immunity 54:1594–1610e11. https://doi.org/10.1016/j.immuni.2021.06.002
Almutairi MM, Sivandzade F, Albekairi TH et al (2021) Neuroinflammation and Its Impact on the Pathogenesis of COVID-19. Front Med 8. https://doi.org/10.3389/fmed.2021.745789
Misra S, Kolappa K, Prasad M et al (2021) Frequency of Neurologic Manifestations in COVID-19. Neurology 97:e2269–e2281. https://doi.org/10.1212/WNL.0000000000012930
Orsucci D, Ienco EC, Nocita G et al (2020) Neurological features of COVID-19 and their treatment: a review. Drugs Context 9. https://doi.org/10.7573/dic.2020-5-1
Berlit P, Bösel J, Gahn G et al (2020) Neurological manifestations of COVID-19” - guideline of the German society of neurology. Neurol Res Pract 2:51. https://doi.org/10.1186/s42466-020-00097-7
Mahdizade Ari M, Mohamadi MH, Shadab Mehr N et al (2022) Neurological manifestations in patients with COVID-19: A systematic review and meta-analysis. J Clin Lab Anal 36:e24403. https://doi.org/10.1002/jcla.24403
Sharma DV, Kumar A, Flora A SJS (2021) Neurological Manifestations in COVID-19 Patients: A Meta-Analysis. ACS Chem Neurosci 12:2776–2797. https://doi.org/10.1021/acschemneuro.1c00353
Rogers JP, Watson CJ, Badenoch J et al (2021) Neurology and neuropsychiatry of COVID-19: a systematic review and meta-analysis of the early literature reveals frequent CNS manifestations and key emerging narratives. J Neurol Neurosurg Psychiatry 92:932–941. https://doi.org/10.1136/jnnp-2021-326405
He Y, Bai X, Zhu T et al (2021) What can the neurological manifestations of COVID-19 tell us: a meta-analysis. J Transl Med 19:363. https://doi.org/10.1186/s12967-021-03039-2
Yassin A, Nawaiseh M, Shaban A et al (2021) Neurological manifestations and complications of coronavirus disease 2019 (COVID-19): a systematic review and meta-analysis. BMC Neurol 21:138. https://doi.org/10.1186/s12883-021-02161-4
Vakili K, Fathi M, Hajiesmaeili M et al (2021) Neurological Symptoms, Comorbidities, and Complications of COVID-19: A Literature Review and Meta-Analysis of Observational Studies. Eur Neurol 84:307–324. https://doi.org/10.1159/000516258
Wang L, Shen Y, Li M et al (2020) Clinical manifestations and evidence of neurological involvement in 2019 novel coronavirus SARS-CoV-2: a systematic review and meta-analysis. J Neurol 267:2777–2789. https://doi.org/10.1007/s00415-020-09974-2
Favas TT, Dev P, Chaurasia RN et al (2020) Neurological manifestations of COVID-19: a systematic review and meta-analysis of proportions. Neurol Sci 41:3437–3470. https://doi.org/10.1007/s10072-020-04801-y
Abdullahi A, Candan SA, Abba MA et al (2020) Neurological and Musculoskeletal Features of COVID-19: A Systematic Review and Meta-Analysis. Front Neurol 11:687. https://doi.org/10.3389/fneur.2020.00687
Lopez-Leon S, Wegman-Ostrosky T, Perelman C et al (2021) More than 50 long-term effects of COVID-19: a systematic review and meta-analysis. Sci Rep 11:16144. https://doi.org/10.1038/s41598-021-95565-8
Khedr EM, Abo-Elfetoh N, Deaf E et al (2021) Surveillance Study of Acute Neurological Manifestations among 439 Egyptian Patients with COVID-19 in Assiut and Aswan University Hospitals. Neuroepidemiology 55:109–118. https://doi.org/10.1159/000513647
Liguori C, Pierantozzi M, Spanetta M, Brain et al (2020) Behav Immun 88:11–16. https://doi.org/10.1016/j.bbi.2020.05.037
Taga A, Lauria G (2022) COVID-19 and the peripheral nervous system. A 2-year review from the pandemic to the vaccine era. J Peripheral Nerv Syst 27:4–30. https://doi.org/10.1111/jns.12482
Aschman T, Stenzel W (2022) COVID-19 associated myopathy. Curr Opin Neurol 35:622. https://doi.org/10.1097/WCO.0000000000001101
García S, Cuatepotzo-Burgos FM, Toledo-Lozano CG et al (2021) Neurological Manifestations and Outcomes in a Retrospective Cohort of Mexican Inpatients with SARS-CoV-2 Pneumonia: Design of a Risk Profile. Healthcare 9. https://doi.org/10.3390/healthcare9111501
Siow I, Lee KS, Zhang JJY et al (2021) Encephalitis as a neurological complication of COVID-19: A systematic review and meta-analysis of incidence, outcomes, and predictors. Eur J Neurol 28:3491–3502. https://doi.org/10.1111/ene.14913
YeMRenYLvT (2020) Encephalitis as a clinical manifestation of COVID-19. Brain Behav Immun 88:945–946. https://doi.org/10.1016/j.bbi.2020.04.017
Bernard-Valnet R, Pizzarotti B, Anichini A et al (2020) Two patients with acute meningoencephalitis concomitant with SARS-CoV-2 infection. Eur J Neurol 27:e43–e44. https://doi.org/10.1111/ene.14298
Moriguchi T, Harii N, Goto J et al (2020) A first case of meningitis/encephalitis associated with SARS-Coronavirus-2. Int J Infect Dis 94:55–58. https://doi.org/10.1016/j.ijid.2020.03.062
Parsons T, Banks S, Bae C et al (2020) COVID-19-associated acute disseminated encephalomyelitis (ADEM). J Neurol 267:2799–2802. https://doi.org/10.1007/s00415-020-09951-9
Vazquez-Guevara D, Badial-Ochoa S, Caceres-Rajo KM, Rodriguez-Leyva I (2021) Catatonic syndrome as the presentation of encephalitis in association with COVID-19. BMJ Case Rep 14:e240550. https://doi.org/10.1136/bcr-2020-240550
Fernández SF, Pérez Sánchez JR, Pérez GH et al (2022) Seizures and COVID-19: Results from the Spanish Society of Neurology’s COVID-19 registry. J Clin Neurosci 101:112–117. https://doi.org/10.1016/j.jocn.2022.05.013
Morassi M, Bagatto D, Cobelli M et al (2020) Stroke in patients with SARS-CoV-2 infection: case series. J Neurol 267:2185–2192. https://doi.org/10.1007/s00415-020-09885-2
Khatana SAM, Groeneveld PW (2020) Health Disparities and the Coronavirus Disease 2019 (COVID-19) Pandemic in the USA. J Gen Intern Med 35:2431–2432. https://doi.org/10.1007/s11606-020-05916-w
Katsoularis I, Fonseca-Rodríguez O, Farrington P et al (2021) Risk of acute myocardial infarction and ischaemic stroke following COVID-19 in Sweden: a self-controlled case series and matched cohort study. The Lancet 398:599–607. https://doi.org/10.1016/S0140-6736(21)00896-5
Katz JM, Libman RB, Wang JJ et al (2020) Cerebrovascular Complications of COVID-19. Stroke 51:e227–e231. https://doi.org/10.1161/STROKEAHA.120.031265
Fridman S, Bres Bullrich M, Jimenez-Ruiz A et al (2020) Stroke risk, phenotypes, and death in COVID-19: Systematic review and newly reported cases. Neurology 95:e3373–e3385. https://doi.org/10.1212/WNL.0000000000010851
Taquet M, Luciano S, Geddes JR, Harrison PJ (2021) Bidirectional associations between COVID-19 and psychiatric disorder: retrospective cohort studies of 62 354 COVID-19 cases in the USA. The Lancet Psychiatry 8:130–140. https://doi.org/10.1016/S2215-0366(20)30462-4
Mahase E (2020) Covid-19: What do we know about “long covid. https://doi.org/10.1136/bmj.m2815 . ? BMJ 370:
Nath A (2020) Long-Haul COVID. Neurology 95:559–560. https://doi.org/10.1212/WNL.0000000000010640
A clinical case definition of post COVID-19 condition by a Delphi consensus, 6 (2021) https://www.who.int/publications-detail-redirect/WHO-2019-nCoV-Post_COVID-19_condition-Clinical_case_definition-2021.1 . Accessed 7 Jan 2023
Castanares-Zapatero D, Chalon P, Kohn L et al (2022) Pathophysiology and mechanism of long COVID: a comprehensive review. Ann Med 54:1473–1487. https://doi.org/10.1080/07853890.2022.2076901
Chen C, Haupert SR, Zimmermann L et al (2022) Global Prevalence of Post-Coronavirus Disease 2019 (COVID-19) Condition or Long COVID: A Meta-Analysis and Systematic Review. J Infect Dis 226:1593–1607. https://doi.org/10.1093/infdis/jiac136
Fernández-de-Las-Peñas C, Palacios-Ceña D, Gómez-Mayordomo V et al (2021) Prevalence of post-COVID-19 symptoms in hospitalized and non-hospitalized COVID-19 survivors: A systematic review and meta-analysis. Eur J Intern Med 92:55–70. https://doi.org/10.1016/j.ejim.2021.06.009
Taquet M, Geddes JR, Husain M et al (2021) 6-month neurological and psychiatric outcomes in 236 379 survivors of COVID-19: a retrospective cohort study using electronic health records. The Lancet Psychiatry 8:416–427. https://doi.org/10.1016/S2215-0366(21)00084-5
Ahmed JO, Ahmad SA, Hassan MN et al (2022) Post COVID-19 neurological complications; a meta-analysis. Ann Med Surg (Lond) 76:103440. https://doi.org/10.1016/j.amsu.2022.103440
Bellan M, Apostolo D, Albè A et al (2022) Determinants of long COVID among adults hospitalized for SARS-CoV-2 infection: A prospective cohort study. Front Immunol 13:1038227. https://doi.org/10.3389/fimmu.2022.1038227
Huang L, Li X, Gu X et al (2022) Health outcomes in people 2 years after surviving hospitalisation with COVID-19: a longitudinal cohort study. The Lancet Respiratory Medicine 10:863–876. https://doi.org/10.1016/S2213-2600(22)00126-6
Ranucci M, Baryshnikova E, Anguissola M et al (2023) The Very Long COVID: Persistence of Symptoms after 12–18 Months from the Onset of Infection and Hospitalization. J Clin Med 12:1915. https://doi.org/10.3390/jcm12051915
Huang L, Yao Q, Gu X et al (2021) 1-year outcomes in hospital survivors with COVID-19: a longitudinal cohort study. The Lancet 398:747–758. https://doi.org/10.1016/S0140-6736(21)01755-4
Søraas A, Kalleberg KT, Dahl JA et al (2021) Persisting symptoms three to eight months after non-hospitalized COVID-19, a prospective cohort study. PLoS ONE 16:e0256142. https://doi.org/10.1371/journal.pone.0256142
Riestra-Ayora J, Yanes-Diaz J, Esteban-Sanchez J et al (2021) Long-term follow-up of olfactory and gustatory dysfunction in COVID-19: 6 months case–control study of health workers. Eur Arch Otorhinolaryngol 278:4831–4837. https://doi.org/10.1007/s00405-021-06764-y
Mattioli F, Stampatori C, Righetti F et al (2021) Neurological and cognitive sequelae of Covid-19: a four month follow-up. J Neurol 268:4422–4428. https://doi.org/10.1007/s00415-021-10579-6
Kubota T, Kuroda N, Sone D (2023) Neuropsychiatric aspects of long COVID: A comprehensive review. J Neuropsychiatry Clin Neurosci 77:84–93. https://doi.org/10.1111/pcn.13508
Desgranges F, Tadini E, Munting A et al (2022) Post–COVID–19 Syndrome in Outpatients: a Cohort Study. J Gen Intern Med 37:1943–1952. https://doi.org/10.1007/s11606-021-07242-1
Chudzik M, Babicki M, Kapusta J et al (2022) Long-COVID Clinical Features and Risk Factors: A Retrospective Analysis of Patients from the STOP-COVID Registry of the PoLoCOV Study. Viruses 14:1755. https://doi.org/10.3390/v14081755
Tsampasian V, Elghazaly H, Chattopadhyay R et al (2023) Risk Factors Associated With Post – COVID-19 Condition: A Systematic Review and Meta-analysis. JAMA Intern Med. https://doi.org/10.1001/jamainternmed.2023.0750
Hedberg P, Granath F, Bruchfeld J et al (2023) Post COVID-19 condition diagnosis: A population‐based cohort study of occurrence, associated factors, and healthcare use by severity of acute infection. J Intern Med 293:246–258. https://doi.org/10.1111/joim.13584
Taquet M, Dercon Q, Luciano S et al (2021) Incidence, co-occurrence, and evolution of long-COVID features: A 6-month retrospective cohort study of 273,618 survivors of COVID-19. PLoS Med 18:e1003773. https://doi.org/10.1371/journal.pmed.1003773
Abu Hamdh B, Nazzal Z (2023) A prospective cohort study assessing the relationship between long-COVID symptom incidence in COVID-19 patients and COVID-19 vaccination. Sci Rep 13:4896. https://doi.org/10.1038/s41598-023-30583-2
Alghamdi SA, Alfares MA, Alsulami RA et al (2022) Post-COVID-19 Syndrome: Incidence, Risk Factor, and the Most Common Persisting Symptoms. Cureus 14:e32058. https://doi.org/10.7759/cureus.32058
Surapaneni KM, Singhal M, Saggu SR et al (2022) A Scoping Review on Long COVID-19: Physiological and Psychological Symptoms Post-Acute, Long-Post and Persistent Post COVID-19. Healthcare 10:2418. https://doi.org/10.3390/healthcare10122418
Fernández-de-las-Peñas C, Pellicer-Valero OJ, Navarro-Pardo E et al (2022) Symptoms Experienced at the Acute Phase of SARS-CoV-2 Infection as Risk Factor of Long-term Post-COVID Symptoms: The LONG-COVID-EXP-CM Multicenter Study. Int J Infect Dis 116:241–244. https://doi.org/10.1016/j.ijid.2022.01.007
Subramanian A, Nirantharakumar K, Hughes S et al (2022) Symptoms and risk factors for long COVID in non-hospitalized adults. Nat Med 28:1706–1714. https://doi.org/10.1038/s41591-022-01909-w
Raj SVA, Jacob A, Ambu V et al (2022) Post COVID-19 clinical manifestations and its risk factors among patients in a Northern District in Kerala, India. J Family Med Prim Care 11:5312–5319. https://doi.org/10.4103/jfmpc.jfmpc_131_22
Guzman-Esquivel J, Mendoza-Hernandez MA, Guzman-Solorzano HP et al (2023) Clinical Characteristics in the Acute Phase of COVID-19 That Predict Long COVID: Tachycardia, Myalgias, Severity, and Use of Antibiotics as Main Risk Factors, While Education and Blood Group B Are Protective. Healthcare 11:197. https://doi.org/10.3390/healthcare11020197
Magnusson K, Kristoffersen DT, Dell’Isola A et al (2022) Post-covid medical complaints following infection with SARS-CoV-2 Omicron vs Delta variants. Nat Commun 13:7363. https://doi.org/10.1038/s41467-022-35240-2
Michelen M, Manoharan L, Elkheir N et al (2021) Characterising long COVID: a living systematic review. BMJ Global Health 6. https://doi.org/10.1136/bmjgh-2021-005427
Alkodaymi MS, Omrani OA, Fawzy NA et al (2022) Prevalence of post-acute COVID-19 syndrome symptoms at different follow-up periods: a systematic review and meta-analysis. Clin Microbiol Infect 28:657–666. https://doi.org/10.1016/j.cmi.2022.01.014
Download references
This research did not receive a specific grant from any organization for the submitted work.
Open access funding provided by Manipal Academy of Higher Education, Manipal
Author information
Authors and affiliations.
Manipal Institute of Virology, Manipal Academy of Higher Education (MAHE), PIN-576104, Manipal, Karnataka, India
Amrita Pattanaik, Sushma Bhandarkar B & Srilatha Marate
Department of Neurovirology, National Institute of Mental Health and Neurosciences (NIMHANS), PIN-560029, Bengaluru, Karnataka, India
Lonika Lodha
You can also search for this author in PubMed Google Scholar
Contributions
Literature search and data analysis was carried out by Amrita Pattanaik, Sushma Bhandarkar, Lonika Lodha, and Srilatha Marate. The manuscript was drafted and critically revised by Amrita Pattanaik.
Corresponding author
Correspondence to Amrita Pattanaik .
Ethics declarations
Conflict of interest.
The authors declare no competing interests.
Ethical approval
None required.
Additional information
Communicated by T. K. Frey
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material
Supplementary Material 1
Rights and permissions.
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Pattanaik, A., Bhandarkar B, S., Lodha, L. et al. SARS-CoV-2 and the nervous system: current perspectives. Arch Virol 168 , 171 (2023). https://doi.org/10.1007/s00705-023-05801-x
Download citation
Received : 09 January 2023
Accepted : 15 April 2023
Published : 01 June 2023
DOI : https://doi.org/10.1007/s00705-023-05801-x
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Neurological impairment
- Neuro COVID
- Post-COVID-19 sequelae
- Find a journal
- Publish with us
- Track your research
Virology Case Study - Answer
A 23 year-old male security guard working in a small department store was stabbed with a needle on a syringe of an intravenous drug addict he was arresting for shoplifting. Two months later he developed general malaise, with nausea and vomiting. He lost his appetite and began to lose weight. When he noticed darkening of the urine and a yellowish color to his sclerae, he came to your office. The patient denied a history of hepatitis or liver disease. He denied drug abuse, and had no history of blood transfusions, surgery or exposure to jaundiced individuals. He never traveled outside the US.
PHYSICAL EXAMINATION
Physical examination revealed a well developed young man in no acute distress. Positive physical findings included: jaundice and a firm, enlarged and tender liver. He had no adenopathy.
Questions and answers:
1. The security guard developed hepatitis 2 months after the incident involving the hypodermic syringe
(a) given the length of time that has elapsed, is it likely that the incident has anything to do with the disease?
Since the incubation time for hepatitis A-E is from about 15 days to about 50, 160, 180, 64, 50 days, respectively, it is quite possible that the incident was relevant to the disease in the security guard.
(b) if the incident had occurred 48 hrs prior to onset of symptoms, would it be likely it had anything to do with the disease?
It would be unlikely, all of these viruses have a much longer incubation time (15 days or more).
2. If one were to acquire hepatitis via a needle stick from the syringe of an intravenous drug addict, what kind(s) of hepatitis would be most likely?
The most likely kinds for parenteral transmission would be hepatitis B, hepatitis C and possibly hepatitis D (which would require the presence of HBV).
Hepatitis A is usually transmitted via the fecal-oral route; there is a viremia with HAV but it is transient (a chronic carrier state does not develop) and although there have been cases of parenteral transmission, they are very rare; so it seems very unlikely that the guard would have gotten HAV from the needle stick.
3. Laboratory tests showed:
HAV Ab(total) Positive HAV Ab(IgM) Positive HBsAg Negative total anti-HBcAg Negative anti-HBsAg Positive HCVAb(total) Negative
(a). How do you interpret the result of the HAV tests?
The security guard has antibodies to HAV and these include IgM antibodies, thus he has had a recent exposure to HAV (since he has IgM antibodies). It is possible that this result is because he has recently been infected with or vaccinated against HAV, further questioning of the guard should clarify this point.
(b). How do you intepret the result of the HBV tests?
Since he has no HBsAg the guard is not a chronic carrier. The guard has anti-HBsAg indicating that he has been either infected with HBV or exposed to the HBsAg present in the vaccine. Thus he has either recovered from HBV infection or he was vaccinated. Since he has no anti-HBcAg it seems probable that this is because he has been vaccinated (there is no HBcAg in the current vaccine). He should be immune to HBV. Again, further questioning of the guard should clarify this point.
(c). Is the security guard likely to be infectious for viral hepatitis?
Causes of viral hepatitis include hepatitis A-E, yellow fever, and it is sometimes seen in infections with Epstein Barr virus. It is possible that there could be more than one virus present. EBV is unlikely since there was no adenopathy (but could be confirmed by further tests). Yellow fever and HEV are unlikely since he had not been out of the country. From the serological data he appears not to have either acute or chronic HBV or HCV. (Because the appearance of anti-HCV may be delayed in patients with acute HCV infection, testing should be repeated if hepatitis C is suspected and initial testing is negative for anti-HCV: anti-HCV is detectable by 5-6 weeks after onset of hepatitis in 80% of patients and by 12 weeks in 90%). The guard is apparently suffering from HAV (by a process of elimination combined with the serology suggesting recent exposure to HAV). Since he has developed IgG antibodies, he is past the period of peak infectivity (furthermore, jaundice with HAV infections typically occurs when patients are past the peak of viral shedding) but there may still be some virus in the feces.
(d). Is he at risk for developing chronic viral hepatitis?
The known forms of chronic viral hepatitis are due to HBV (the guard has been vaccinated and has protective antibody), HCV (the serological results were negative, so he does not seem to have HCV but should be retested, if exposed in the future he has no protective antibodies, and there is no vaccine at the moment) and HDV (no risk if the guard has protective antibody against HBV). So the guard is probably not in imminent danger of developing chronic viral hepatitis (unless he turns out in a future test to have HCV) and is not currently a chronic carrier of viral hepatitis.
(e). Should he be tested for HDV?
Since he does not have HBV, he cannot be infected with HDV, so testing is unnecessary.
Note: since the guard most probably did not get the HAV from the needle stick, other sources of infection should be considered, such as via a child in the family, eating of raw seafood such as oysters, possibly eating of contaminated uncooked food etc.
Note: there are other causes of hepatitis, such as CMV, toxoplasmosis, leptospirosis, secondary syphilis, exposure to toxic chemicals (including alcohol), toxic drug reactions, etc. If the guard had been vaccinated recently against HAV, alternative explanations should be explored.
Comment: The risk of acquiring a blood borne viral infection from an infected individual via a needle stick varies widely from pathogen to pathogen, according to the pathogen, the degree of viremia, the details of the stick, etc. but very broadly speaking risk values are:
HBV in the order of 30%; HCV in the order of 3%; HIV in the order of 0.3% Return to the Department of Microbiology and Immunology Site Guide This page copyright 1998, The Board of Trustees of the University of South Carolina This page last changed on Wednesday, May 28, 2003 Page maintained by Richard Hunt URL: http://www.med.sc.edu:85/mhunt/case-answ.htm Please report any problems to [email protected]

- Academic Programs
Molecular Virology
Download our Graduate Student Handbook
Requirements
The requirements for a PhD degree in Molecular Virology include course work, participation in formal and informal seminars, a research qualifying examination, and the PhD thesis.
In addition to the core curriculum, students are required to complete a minimum of sixteen additional credit hours of advanced course work, including Introduction to Virology and Virus-Host Interactions. Recent examples of specialized, elective courses relevant to the program include: Mechanisms of Drug Resistance, Molecular Genetics of Cancer, Molecular Biology of RNA, Transcription and Gene Regulation, Immunology of Infectious Disease, and HIV & AIDS: Research and Care.
Formal and Informal Seminars
Students also participate in formal and informal seminars. Each year, students present a seminar on their own research to members of the department. Prominent outside speakers are invited to present current research through the weekly Molecular Biology & Microbiology seminar program, the monthly Lester O. Krampitz Seminars and Cleveland Virology Group meetings, and numerous seminars in other departments.
Qualifying Exam
The qualifying exam for advancement to candidacy for the PhD degree focuses on the student's own research and consists of two parts. In the first part, an original description of the student's research project is prepared in the form of a grant application. In the second part, the student defends the written proposal before the pre-thesis committee in an oral examination. Preparation for this exam provides experience in developing and formulating original research ideas and assessing their feasibility. At the time of the qualifying examination, a student is expected to be conversant with relevant experimental techniques and to be familiar with the scientific literature pertinent to the proposed project. Generally, the qualifying exam is completed by the middle of the third year of study.
For the PhD thesis, the student will carry out an original research project in the advisor's laboratory. Early in the second year, a pre-thesis committee, consisting of the thesis advisor and additional faculty members, will be chosen to advise the student on the academic program and the dissertation. This committee provides scientific expertise, offers support in overcoming research difficulties, and evaluates progress towards the PhD degree.
- Open access
- Published: 22 April 2024
Artificial intelligence and medical education: application in classroom instruction and student assessment using a pharmacology & therapeutics case study
- Kannan Sridharan 1 &
- Reginald P. Sequeira 1
BMC Medical Education volume 24 , Article number: 431 ( 2024 ) Cite this article
454 Accesses
1 Altmetric
Metrics details
Artificial intelligence (AI) tools are designed to create or generate content from their trained parameters using an online conversational interface. AI has opened new avenues in redefining the role boundaries of teachers and learners and has the potential to impact the teaching-learning process.
In this descriptive proof-of- concept cross-sectional study we have explored the application of three generative AI tools on drug treatment of hypertension theme to generate: (1) specific learning outcomes (SLOs); (2) test items (MCQs- A type and case cluster; SAQs; OSPE); (3) test standard-setting parameters for medical students.
Analysis of AI-generated output showed profound homology but divergence in quality and responsiveness to refining search queries. The SLOs identified key domains of antihypertensive pharmacology and therapeutics relevant to stages of the medical program, stated with appropriate action verbs as per Bloom’s taxonomy. Test items often had clinical vignettes aligned with the key domain stated in search queries. Some test items related to A-type MCQs had construction defects, multiple correct answers, and dubious appropriateness to the learner’s stage. ChatGPT generated explanations for test items, this enhancing usefulness to support self-study by learners. Integrated case-cluster items had focused clinical case description vignettes, integration across disciplines, and targeted higher levels of competencies. The response of AI tools on standard-setting varied. Individual questions for each SAQ clinical scenario were mostly open-ended. The AI-generated OSPE test items were appropriate for the learner’s stage and identified relevant pharmacotherapeutic issues. The model answers supplied for both SAQs and OSPEs can aid course instructors in planning classroom lessons, identifying suitable instructional methods, establishing rubrics for grading, and for learners as a study guide. Key lessons learnt for improving AI-generated test item quality are outlined.
Conclusions
AI tools are useful adjuncts to plan instructional methods, identify themes for test blueprinting, generate test items, and guide test standard-setting appropriate to learners’ stage in the medical program. However, experts need to review the content validity of AI-generated output. We expect AIs to influence the medical education landscape to empower learners, and to align competencies with curriculum implementation. AI literacy is an essential competency for health professionals.
Peer Review reports
Artificial intelligence (AI) has great potential to revolutionize the field of medical education from curricular conception to assessment [ 1 ]. AIs used in medical education are mostly generative AI large language models that were developed and validated based on billions to trillions of parameters [ 2 ]. AIs hold promise in the incorporation of history-taking, assessment, diagnosis, and management of various disorders [ 3 ]. While applications of AIs in undergraduate medical training are being explored, huge ethical challenges remain in terms of data collection, maintaining anonymity, consent, and ownership of the provided data [ 4 ]. AIs hold a promising role amongst learners because they can deliver a personalized learning experience by tracking their progress and providing real-time feedback, thereby enhancing their understanding in the areas they are finding difficult [ 5 ]. Consequently, a recent survey has shown that medical students have expressed their interest in acquiring competencies related to the use of AIs in healthcare during their undergraduate medical training [ 6 ].
Pharmacology and Therapeutics (P & T) is a core discipline embedded in the undergraduate medical curriculum, mostly in the pre-clerkship phase. However, the application of therapeutic principles forms one of the key learning objectives during the clerkship phase of the undergraduate medical career. Student assessment in pharmacology & therapeutics (P&T) is with test items such as multiple-choice questions (MCQs), integrated case cluster questions, short answer questions (SAQs), and objective structured practical examination (OSPE) in the undergraduate medical curriculum. It has been argued that AIs possess the ability to communicate an idea more creatively than humans [ 7 ]. It is imperative that with access to billions of trillions of datasets the AI platforms hold promise in playing a crucial role in the conception of various test items related to any of the disciplines in the undergraduate medical curriculum. Additionally, AIs provide an optimized curriculum for a program/course/topic addressing multidimensional problems [ 8 ], although robust evidence for this claim is lacking.
The existing literature has evaluated the knowledge, attitude, and perceptions of adopting AI in medical education. Integration of AIs in medical education is the need of the hour in all health professional education. However, the academic medical fraternity facing challenges in the incorporation of AIs in the medical curriculum due to factors such as inadequate grounding in data analytics, lack of high-quality firm evidence favoring the utility of AIs in medical education, and lack of funding [ 9 ]. Open-access AI platforms are available free to users without any restrictions. Hence, as a proof-of-concept, we chose to explore the utility of three AI platforms to identify specific learning objectives (SLOs) related to pharmacology discipline in the management of hypertension for medical students at different stages of their medical training.
Study design and ethics
The present study is observational, cross-sectional in design, conducted in the Department of Pharmacology & Therapeutics, College of Medicine and Medical Sciences, Arabian Gulf University, Kingdom of Bahrain, between April and August 2023. Ethical Committee approval was not sought given the nature of this study that neither had any interaction with humans, nor collection of any personal data was involved.
Study procedure
We conducted the present study in May-June 2023 with the Poe© chatbot interface created by Quora© that provides access to the following three AI platforms:
Sage Poe [ 10 ]: A generative AI search engine developed by Anthropic © that conceives a response based on the written input provided. Quora has renamed Sage Poe as Assistant © from July 2023 onwards.
Claude-Instant [ 11 ]: A retrieval-based AI search engine developed by Anthropic © that collates a response based on pre-written responses amongst the existing databases.
ChatGPT version 3.5 [ 12 ]: A generative architecture-based AI search engine developed by OpenAI © trained on large and diverse datasets.
We queried the chatbots to generate SLOs, A-type MCQs, integrated case cluster MCQs, integrated SAQs, and OSPE test items in the domain of systemic hypertension related to the P&T discipline. Separate prompts were used to generate outputs for pre-clerkship (preclinical) phase students, and at the time of graduation (before starting residency programs). Additionally, we have also evaluated the ability of these AI platforms to estimate the proportion of students correctly answering these test items. We used the following queries for each of these objectives:
Specific learning objectives
Can you generate specific learning objectives in the pharmacology discipline relevant to undergraduate medical students during their pre-clerkship phase related to anti-hypertensive drugs?
Can you generate specific learning objectives in the pharmacology discipline relevant to undergraduate medical students at the time of graduation related to anti-hypertensive drugs?
A-type MCQs
In the initial query used for A-type of item, we specified the domains (such as the mechanism of action, pharmacokinetics, adverse reactions, and indications) so that a sample of test items generated without any theme-related clutter, shown below:
Write 20 single best answer MCQs with 5 choices related to anti-hypertensive drugs for undergraduate medical students during the pre-clerkship phase of which 5 MCQs should be related to mechanism of action, 5 MCQs related to pharmacokinetics, 5 MCQs related to adverse reactions, and 5 MCQs should be related to indications.
The MCQs generated with the above search query were not based on clinical vignettes. We queried again to generate MCQs using clinical vignettes specifically because most medical schools have adopted problem-based learning (PBL) in their medical curriculum.
Write 20 single best answer MCQs with 5 choices related to anti-hypertensive drugs for undergraduate medical students during the pre-clerkship phase using a clinical vignette for each MCQ of which 5 MCQs should be related to the mechanism of action, 5 MCQs related to pharmacokinetics, 5 MCQs related to adverse reactions, and 5 MCQs should be related to indications.
We attempted to explore whether AI platforms can provide useful guidance on standard-setting. Hence, we used the following search query.
Can you do a simulation with 100 undergraduate medical students to take the above questions and let me know what percentage of students got each MCQ correct?
Integrated case cluster MCQs
Write 20 integrated case cluster MCQs with 2 questions in each cluster with 5 choices for undergraduate medical students during the pre-clerkship phase integrating pharmacology and physiology related to systemic hypertension with a case vignette.
Write 20 integrated case cluster MCQs with 2 questions in each cluster with 5 choices for undergraduate medical students during the pre-clerkship phase integrating pharmacology and physiology related to systemic hypertension with a case vignette. Please do not include ‘none of the above’ as the choice. (This modified search query was used because test items with ‘None of the above’ option were generated with the previous search query).
Write 20 integrated case cluster MCQs with 2 questions in each cluster with 5 choices for undergraduate medical students at the time of graduation integrating pharmacology and physiology related to systemic hypertension with a case vignette.
Integrated short answer questions
Write a short answer question scenario with difficult questions based on the theme of a newly diagnosed hypertensive patient for undergraduate medical students with the main objectives related to the physiology of blood pressure regulation, risk factors for systemic hypertension, pathophysiology of systemic hypertension, pathological changes in the systemic blood vessels in hypertension, pharmacological management, and non-pharmacological treatment of systemic hypertension.
Write a short answer question scenario with moderately difficult questions based on the theme of a newly diagnosed hypertensive patient for undergraduate medical students with the main objectives related to the physiology of blood pressure regulation, risk factors for systemic hypertension, pathophysiology of systemic hypertension, pathological changes in the systemic blood vessels in hypertension, pharmacological management, and non-pharmacological treatment of systemic hypertension.
Write a short answer question scenario with questions based on the theme of a newly diagnosed hypertensive patient for undergraduate medical students at the time of graduation with the main objectives related to the physiology of blood pressure regulation, risk factors for systemic hypertension, pathophysiology of systemic hypertension, pathological changes in the systemic blood vessels in hypertension, pharmacological management, and non-pharmacological treatment of systemic hypertension.
Can you generate 5 OSPE pharmacology and therapeutics prescription writing exercises for the assessment of undergraduate medical students at the time of graduation related to anti-hypertensive drugs?
Can you generate 5 OSPE pharmacology and therapeutics prescription writing exercises containing appropriate instructions for the patients for the assessment of undergraduate medical students during their pre-clerkship phase related to anti-hypertensive drugs?
Can you generate 5 OSPE pharmacology and therapeutics prescription writing exercises containing appropriate instructions for the patients for the assessment of undergraduate medical students at the time of graduation related to anti-hypertensive drugs?
Both authors independently evaluated the AI-generated outputs, and a consensus was reached. We cross-checked the veracity of answers suggested by AIs as per the Joint National Commission Guidelines (JNC-8) and Goodman and Gilman’s The Pharmacological Basis of Therapeutics (2023), a reference textbook [ 13 , 14 ]. Errors in the A-type MCQs were categorized as item construction defects, multiple correct answers, and uncertain appropriateness to the learner’s level. Test items in the integrated case cluster MCQs, SAQs and OSPEs were evaluated with the Preliminary Conceptual Framework for Establishing Content Validity of AI-Generated Test Items based on the following domains: technical accuracy, comprehensiveness, education level, and lack of construction defects (Table 1 ). The responses were categorized as complete and deficient for each domain.
The pre-clerkship phase SLOs identified by Sage Poe, Claude-Instant, and ChatGPT are listed in the electronic supplementary materials 1 – 3 , respectively. In general, a broad homology in SLOs generated by the three AI platforms was observed. All AI platforms identified appropriate action verbs as per Bloom’s taxonomy to state the SLO; action verbs such as describe, explain, recognize, discuss, identify, recommend, and interpret are used to state the learning outcome. The specific, measurable, achievable, relevant, time-bound (SMART) SLOs generated by each AI platform slightly varied. All key domains of antihypertensive pharmacology to be achieved during the pre-clerkship (pre-clinical) years were relevant for graduating doctors. The SLOs addressed current JNC Treatment Guidelines recommended classes of antihypertensive drugs, the mechanism of action, pharmacokinetics, adverse effects, indications/contraindications, dosage adjustments, monitoring therapy, and principles of monotherapy and combination therapy.
The SLOs to be achieved by undergraduate medical students at the time of graduation identified by Sage Poe, Claude-Instant, and ChatGPT listed in electronic supplementary materials 4 – 6 , respectively. The identified SLOs emphasize the application of pharmacology knowledge within a clinical context, focusing on competencies needed to function independently in early residency stages. These SLOs go beyond knowledge recall and mechanisms of action to encompass competencies related to clinical problem-solving, rational prescribing, and holistic patient management. The SLOs generated require higher cognitive ability of the learner: action verbs such as demonstrate, apply, evaluate, analyze, develop, justify, recommend, interpret, manage, adjust, educate, refer, design, initiate & titrate were frequently used.
The MCQs for the pre-clerkship phase identified by Sage Poe, Claude-Instant, and ChatGPT listed in the electronic supplementary materials 7 – 9 , respectively, and those identified with the search query based on the clinical vignette in electronic supplementary materials ( 10 – 12 ).
All MCQs generated by the AIs in each of the four domains specified [mechanism of action (MOA); pharmacokinetics; adverse drug reactions (ADRs), and indications for antihypertensive drugs] are quality test items with potential content validity. The test items on MOA generated by Sage Poe included themes such as renin-angiotensin-aldosterone (RAAS) system, beta-adrenergic blockers (BB), calcium channel blockers (CCB), potassium channel openers, and centrally acting antihypertensives; on pharmacokinetics included high oral bioavailability/metabolism in liver [angiotensin receptor blocker (ARB)-losartan], long half-life and renal elimination [angiotensin converting enzyme inhibitors (ACEI)-lisinopril], metabolism by both liver and kidney (beta-blocker (BB)-metoprolol], rapid onset- short duration of action (direct vasodilator-hydralazine), and long-acting transdermal drug delivery (centrally acting-clonidine). Regarding the ADR theme, dry cough, angioedema, and hyperkalemia by ACEIs in susceptible patients, reflex tachycardia by CCB/amlodipine, and orthostatic hypotension by CCB/verapamil addressed. Clinical indications included the drug of choice for hypertensive patients with concomitant comorbidity such as diabetics (ACEI-lisinopril), heart failure and low ejection fraction (BB-carvedilol), hypertensive urgency/emergency (alpha cum beta receptor blocker-labetalol), stroke in patients with history recurrent stroke or transient ischemic attack (ARB-losartan), and preeclampsia (methyldopa).
Almost similar themes under each domain were identified by the Claude-Instant AI platform with few notable exceptions: hydrochlorothiazide (instead of clonidine) in MOA and pharmacokinetics domains, respectively; under the ADR domain ankle edema/ amlodipine, sexual dysfunction and fatigue in male due to alpha-1 receptor blocker; under clinical indications the best initial monotherapy for clinical scenarios such as a 55-year old male with Stage-2 hypertension; a 75-year-old man Stage 1 hypertension; a 35-year-old man with Stage I hypertension working on night shifts; and a 40-year-old man with stage 1 hypertension and hyperlipidemia.
As with Claude-Instant AI, ChatGPT-generated test items on MOA were mostly similar. However, under the pharmacokinetic domain, immediate- and extended-release metoprolol, the effect of food to enhance the oral bioavailability of ramipril, and the highest oral bioavailability of amlodipine compared to other commonly used antihypertensives were the themes identified. Whereas the other ADR themes remained similar, constipation due to verapamil was a new theme addressed. Notably, in this test item, amlodipine was an option that increased the difficulty of this test item because amlodipine therapy is also associated with constipation, albeit to a lesser extent, compared to verapamil. In the clinical indication domain, the case description asking “most commonly used in the treatment of hypertension and heart failure” is controversial because the options listed included losartan, ramipril, and hydrochlorothiazide but the suggested correct answer was ramipril. This is a good example to stress the importance of vetting the AI-generated MCQ by experts for content validity and to assure robust psychometrics. The MCQ on the most used drug in the treatment of “hypertension and diabetic nephropathy” is more explicit as opposed to “hypertension and diabetes” by Claude-Instant because the therapeutic concept of reducing or delaying nephropathy must be distinguished from prevention of nephropathy, although either an ACEI or ARB is the drug of choice for both indications.
It is important to align student assessment to the curriculum; in the PBL curriculum, MCQs with a clinical vignette are preferred. The modification of the query specifying the search to generate MCQs with a clinical vignette on domains specified previously gave appropriate output by all three AI platforms evaluated (Sage Poe; Claude- Instant; Chat GPT). The scenarios generated had a good clinical fidelity and educational fit for the pre-clerkship student perspective.
The errors observed with AI outputs on the A-type MCQs are summarized in Table 2 . No significant pattern was observed except that Claude-Instant© generated test items in a stereotyped format such as the same choices for all test items related to pharmacokinetics and indications, and all the test items in the ADR domain are linked to the mechanisms of action of drugs. This illustrates the importance of reviewing AI-generated test items by content experts for content validity to ensure alignment with evidence-based medicine and up-to-date treatment guidelines.
The test items generated by ChatGPT had the advantage of explanations supplied rendering these more useful for learners to support self-study. The following examples illustrate this assertion: “ A patient with hypertension is started on a medication that works by blocking beta-1 receptors in the heart (metoprolol)”. Metoprolol is a beta blocker that works by blocking beta-1 receptors in the heart, which reduces heart rate and cardiac output, resulting in a decrease in blood pressure. However, this explanation is incomplete because there is no mention of other less important mechanisms, of beta receptor blockers on renin release. Also, these MCQs were mostly recall type: Which of the following medications is known to have a significant first-pass effect? The explanation reads: propranolol is known to have a significant first pass-effect, meaning that a large portion of the drug is metabolized by the liver before it reaches systemic circulation. Losartan, amlodipine, ramipril, and hydrochlorothiazide do not have significant first-pass effect. However, it is also important to extend the explanation further by stating that the first-pass effect of propranolol does not lead to total loss of pharmacological activity because the metabolite hydroxy propranolol also has potent beta-blocking activity. Another MCQ test item had a construction defect: “A patient with hypertension is started on a medication that can cause photosensitivity. Which of the following medications is most likely responsible?” Options included: losartan, amlodipine, ramipril, hydrochlorothiazide, hydrochlorothiazide/triamterene. The explanation states that hydrochlorothiazide (HCTZ) can cause photosensitivity, which is increased sensitivity to sunlight that can cause skin rashes and sunburns. Losartan, amlodipine, ramipril, and HCTZ/triamterene are less likely to cause photosensitivity. However, it is irrational to claim that HCTZ/triamterene, one of the wrong options, is less likely to cause photosensitivity. The difficulty index of such test items is likely unacceptable in high-stake tests to make equitable psychometric decisions.
The responses from Sage Poe, Claude-Instant, and ChatGPT on guidance for standard-setting methods are listed in the electronic supplementary materials 13 – 15 . Sage Poe offered this information with the caveat that “the results of this simulation were purely hypothetical and do not reflect the actual performance of any real students”. The simulation estimates of examinees correctly answering test items ranged from 60 to 90% for the mechanism of action, 65–85% for pharmacokinetics, 60–80% for adverse reactions, and 75–90% for clinical indications of antihypertensives.
In contrast, Claude-Instant admitted that “Performing a full simulation with 100 medical students is beyond the scope of what I can do as an AI assistant. I can generate sample MCQ questions and answers, but I do not have access to a population of medical students to administer the questions. A true simulation that administers questions to real students and analyzes the results would require access to a database of medical students, a platform to deliver the questions, and code to analyze the responses. As an AI assistant, I lack these capabilities. While I cannot perform a full simulation, here are some considerations for administering MCQ questions to medical students: provide details on the purpose and instructions for the test to ensure students understand what is expected out of them; consider including some easier questions in addition to more challenging ones to boost students’ confidence and motivation; avoid ambiguous wording and ensure each question has only one unambiguously correct answer option; when analyzing results, look for questions factors that may have impacted performance, like unclear wording or answer options that are too similar. Focusing on weak questions can help improve future tests; compare performance across different student groups to identify areas where more teaching may be needed; provide detailed answer explanations and review sessions after the test to help students learn from any errors and reinforce correct concepts”. However, ChatGPT offered the simulation result estimates for the 20 MCQ on antihypertensive drugs: mechanism of action ranged from 67 to 92%, pharmacokinetics 63–86%, adverse effects 65–82%, and clinical indications 64–89%. Furthermore, it also stated that “Overall, the performance of the students was quite good, with most questions having a response rate of over 70%. However, there were some questions where the projected correct response rate was rather low, such as Question #5 (Mechanism of Action of Hydrochlorothiazide; see Electronic Supplementary Material 12 ) and Question 18 (Indications for Verapamil; see Electronic Supplementary Material 10 ). This may suggest areas where students need more focused education or review.”
We asked AI assistants to generate 20 integrated case cluster MCQs with 2 test items in each cluster with five options for undergraduate medical students in the pre-clerkship phase integrating pharmacology and physiology related to systemic hypertension with a case vignette and the responses by Sage Poe, Claude-Instant, and ChatGPT are listed in the electronic supplementary materials ( 16 – 18 ). In all instances, the test items generated had focused case descriptions in the form of a clinical vignette, and horizontal integration across the pathophysiology of hypertension and pharmacology of antihypertensive drugs. These test items mostly targeted the ‘knows (knowledge)’ or ‘knows how (competence)’ level on Miller’s pyramid and are suitable for assessing the clinical competence of pre-clerkship medical students, especially in an integrated PBL curriculum. Both the AI assistants generated excellent clinical vignettes and themes; however, most of the cluster MCQs by ChatGPT had “None of the above” as an option, which is often considered a test item construction flaw. Notwithstanding these limitations, case cluster integrated test items are valuable for learners to integrate their knowledge of different basic medical sciences and their application to clinical sciences. This integrated approach can be used for both instructional and student assessment purposes to make the course more meaningful. Indeed, one of the basic tenets of PBL is curriculum integration.
In the next query, we asked AI assistants to write integrated case cluster MCQs with 2 test items in each cluster with 5 options for undergraduate medical students at the time of graduation integrating pharmacology and physiology of hypertension with a case vignette. The responses are listed in the electronic supplementary materials 19 – 21 . Claude-Instant has provided only partial responses to both the queries. Overall, the information provided in case vignettes requires the learner to interpret the clinical information and make therapeutic decisions. Such test items are appropriate for assessing the minimum competency of medical school graduates to start internship or residency programs; these MCQs are generally comparable to test items used in USMLE Part 2. The evaluation of the integrated case cluster test items provided by all the AI platforms is summarized in Table 3 . In general, the AI outputs had at least one construction defect, and did not include all the core sub-topics to a sufficient degree.
When asked to simulate the above-generated test items to 100 undergraduate medical students at the time of graduation and provide a percentage of students who are likely to correctly answer each item, the AI assistant responded:
“ As an AI language model, I cannot simulate the responses of 100 undergraduate medical students likely to obtain the correct answer for each question. However, I can provide some general information about the difficulty level of each question based on the complexity of the concepts being tested and the level of integration between pharmacology and physiology ”.
AI assistant went on to clarify that questions that require knowledge of basic pharmacology principles, such as the mechanism of action of specific drugs, are likely to be easier for students to answer correctly. Test items that require an understanding of the physiological mechanisms underlying hypertension and correlating with symptoms are likely to be more challenging for students. The AI assistant sorted these test items into two categories accordingly. Overall, the difficulty level of the test item is based on the level of integration between pharmacology and pathophysiology. Test items that require an understanding of both pharmacological and physiological mechanisms are likely to be more challenging for students requiring a strong foundation in both pharmacology and physiology concepts to be able to correctly answer integrated case-cluster MCQs.
Short answer questions
The responses to a search query on generating SAQs appropriate to the pre-clerkship phase Sage Poe, Claude-Instant, and ChatGPT generated items are listed in the electronic supplementary materials 22 – 24 for difficult questions and 25–27 for moderately difficult questions.
It is apparent from these case vignette descriptions that the short answer question format varied. Accordingly, the scope for asking individual questions for each scenario is open-ended. In all instances, model answers are supplied which are helpful for the course instructor to plan classroom lessons, identify appropriate instructional methods, and establish rubrics for grading the answer scripts, and as a study guide for students.
We then wanted to see to what extent AI can differentiate the difficulty of the SAQ by replacing the search term “difficult” with “moderately difficult” in the above search prompt: the changes in the revised case scenarios are substantial. Perhaps the context of learning and practice (and the level of the student in the MD/medical program) may determine the difficulty level of SAQ generated. It is worth noting that on changing the search from cardiology to internal medicine rotation in Sage Poe the case description also changed. Thus, it is essential to select an appropriate AI assistant, perhaps by trial and error, to generate quality SAQs. Most of the individual questions tested stand-alone knowledge and did not require students to demonstrate integration.
The responses of Sage Poe, Claude-Instant, and ChatGPT for the search query to generate SAQs at the time of graduation are listed in the electronic supplementary materials 28 – 30 . It is interesting to note how AI assistants considered the stage of the learner while generating the SAQ. The response by Sage Poe is illustrative for comparison. “You are a newly graduated medical student who is working in a hospital” versus “You are a medical student in your pre-clerkship.”
Some questions were retained, deleted, or modified to align with competency appropriate to the context (Electronic Supplementary Materials 28 – 30 ). Overall, the test items at both levels from all AI platforms were technically accurate and thorough addressing the topics related to different disciplines (Table 3 ). The differences in learning objective transition are summarized in Table 4 . A comparison of learning objectives revealed that almost all objectives remained the same except for a few (Table 5 ).
A similar trend was apparent with test items generated by other AI assistants, such as ChatGPT. The contrasting differences in questions are illustrated by the vertical integration of basic sciences and clinical sciences (Table 6 ).
Taken together, these in-depth qualitative comparisons suggest that AI assistants such as Sage Poe and ChatGPT consider the learner’s stage of training in designing test items, learning outcomes, and answers expected from the examinee. It is critical to state the search query explicitly to generate quality output by AI assistants.
The OSPE test items generated by Claude-Instant and ChatGPT appropriate to the pre-clerkship phase (without mentioning “appropriate instructions for the patients”) are listed in the electronic supplementary materials 31 and 32 and with patient instructions on the electronic supplementary materials 33 and 34 . For reasons unknown, Sage Poe did not provide any response to this search query.
The five OSPE items generated were suitable to assess the prescription writing competency of pre-clerkship medical students. The clinical scenarios identified by the three AI platforms were comparable; these scenarios include patients with hypertension and impaired glucose tolerance in a 65-year-old male, hypertension with chronic kidney disease (CKD) in a 55-year-old woman, resistant hypertension with obstructive sleep apnea in a 45-year-old man, and gestational hypertension at 32 weeks in a 35-year-old (Claude-Instant AI). Incorporating appropriate instructions facilitates the learner’s ability to educate patients and maximize safe and effective therapy. The OSPE item required students to write a prescription with guidance to start conservatively, choose an appropriate antihypertensive drug class (drug) based on the patients’ profile, specifying drug name, dose, dosing frequency, drug quantity to be dispensed, patient name, date, refill, and caution as appropriate, in addition to prescribers’ name, signature, and license number. In contrast, ChatGPT identified clinical scenarios to include patients with hypertension and CKD, hypertension and bronchial asthma, gestational diabetes, hypertension and heart failure, and hypertension and gout (ChatGPT). Guidance for dosage titration, warnings to be aware, safety monitoring, and frequency of follow-up and dose adjustment. These test items are designed to assess learners’ knowledge of P & T of antihypertensives, as well as their ability to provide appropriate instructions to patients. These clinical scenarios for writing prescriptions assess students’ ability to choose an appropriate drug class, write prescriptions with proper labeling and dosing, reflect drug safety profiles, and risk factors, and make modifications to meet the requirements of special populations. The prescription is required to state the drug name, dose, dosing frequency, patient name, date, refills, and cautions or instructions as needed. A conservative starting dose, once or twice daily dosing frequency based on the drug, and instructions to titrate the dose slowly if required.
The responses from Claude-Instant and ChatGPT for the search query related to generating OSPE test items at the time of graduation are listed in electronic supplementary materials 35 and 36 . In contrast to the pre-clerkship phase, OSPEs generated for graduating doctors’ competence assessed more advanced drug therapy comprehension. For example, writing a prescription for:
(1) A 65-year- old male with resistant hypertension and CKD stage 3 to optimize antihypertensive regimen required the answer to include starting ACEI and diuretic, titrating the dosage over two weeks, considering adding spironolactone or substituting ACEI with an ARB, and need to closely monitor serum electrolytes and kidney function closely.
(2) A 55-year-old woman with hypertension and paroxysmal arrhythmia required the answer to include switching ACEI to ARB due to cough, adding a CCB or beta blocker for rate control needs, and adjusting the dosage slowly and monitoring for side effects.
(3) A 45-year-old man with masked hypertension and obstructive sleep apnea require adding a centrally acting antihypertensive at bedtime and increasing dosage as needed based on home blood pressure monitoring and refer to CPAP if not already using one.
(4) A 75-year-old woman with isolated systolic hypertension and autonomic dysfunction to require stopping diuretic and switching to an alpha blocker, upward dosage adjustment and combining with other antihypertensives as needed based on postural blood pressure changes and symptoms.
(5) A 35-year-old pregnant woman with preeclampsia at 29 weeks require doubling methyldopa dose and consider adding labetalol or nifedipine based on severity and educate on signs of worsening and to follow-up immediately for any concerning symptoms.
These case scenarios are designed to assess the ability of the learner to comprehend the complexity of antihypertensive regimens, make evidence-based regimen adjustments, prescribe multidrug combinations based on therapeutic response and tolerability, monitor complex patients for complications, and educate patients about warning signs and follow-up.
A similar output was provided by ChatGPT, with clinical scenarios such as prescribing for patients with hypertension and myocardial infarction; hypertension and chronic obstructive pulmonary airway disease (COPD); hypertension and a history of angina; hypertension and a history of stroke, and hypertension and advanced renal failure. In these cases, wherever appropriate, pharmacotherapeutic issues like taking ramipril after food to reduce side effects such as giddiness; selection of the most appropriate beta-blocker such as nebivolol in patients with COPD comorbidity; the importance of taking amlodipine at the same time every day with or without food; preference for telmisartan among other ARBs in stroke; choosing furosemide in patients with hypertension and edema and taking the medication with food to reduce the risk of gastrointestinal adverse effect are stressed.
The AI outputs on OSPE test times were observed to be technically accurate, thorough in addressing core sub-topics suitable for the learner’s level and did not have any construction defects (Table 3 ). Both AIs provided the model answers with explanatory notes. This facilitates the use of such OSPEs for self-assessment by learners for formative assessment purposes. The detailed instructions are helpful in creating optimized therapy regimens, and designing evidence-based regimens, to provide appropriate instructions to patients with complex medical histories. One can rely on multiple AI sources to identify, shortlist required case scenarios, and OSPE items, and seek guidance on expected model answers with explanations. The model answer guidance for antihypertensive drug classes is more appropriate (rather than a specific drug of a given class) from a teaching/learning perspective. We believe that these scenarios can be refined further by providing a focused case history along with relevant clinical and laboratory data to enhance clinical fidelity and bring a closer fit to the competency framework.
In the present study, AI tools have generated SLOs that comply with the current principles of medical education [ 15 ]. AI tools are valuable in constructing SLOs and so are especially useful for medical fraternities where training in medical education is perceived as inadequate, more so in the early stages of their academic career. Data suggests that only a third of academics in medical schools have formal training in medical education [ 16 ] which is a limitation. Thus, the credibility of alternatives, such as the AIs, is evaluated to generate appropriate course learning outcomes.
We observed that the AI platforms in the present study generated quality test items suitable for different types of assessment purposes. The AI-generated outputs were similar with minor variation. We have used generative AIs in the present study that could generate new content from their training dataset [ 17 ]. Problem-based and interactive learning approaches are referred to as “bottom-up” where learners obtain first-hand experience in solving the cases first and then indulge in discussion with the educators to refine their understanding and critical thinking skills [ 18 ]. We suggest that AI tools can be useful for this approach for imparting the core knowledge and skills related to Pharmacology and Therapeutics to undergraduate medical students. A recent scoping review evaluating the barriers to writing quality test items based on 13 studies has concluded that motivation, time constraints, and scheduling were the most common [ 19 ]. AI tools can be valuable considering the quick generation of quality test items and time management. However, as observed in the present study, the AI-generated test items nevertheless require scrutiny by faculty members for content validity. Moreover, it is important to train faculty in AI technology-assisted teaching and learning. The General Medical Council recommends taking every opportunity to raise the profile of teaching in medical schools [ 20 ]. Hence, both the academic faculty and the institution must consider investing resources in AI training to ensure appropriate use of the technology [ 21 ].
The AI outputs assessed in the present study had errors, particularly with A-type MCQs. One notable observation was that often the AI tools were unable to differentiate the differences between ACEIs and ARBs. AI platforms access several structured and unstructured data, in addition to images, audio, and videos. Hence, the AI platforms can commit errors due to extracting details from unauthenticated sources [ 22 ] created a framework identifying 28 factors for reconstructing the path of AI failures and for determining corrective actions. This is an area of interest for AI technical experts to explore. Also, this further iterates the need for human examination of test items before using them for assessment purposes.
There are concerns that AIs can memorize and provide answers from their training dataset, which they are not supposed to do [ 23 ]. Hence, the use of AIs-generated test items for summative examinations is debatable. It is essential to ensure and enhance the security features of AI tools to reduce or eliminate cross-contamination of test items. Researchers have emphasized that AI tools will only reach their potential if developers and users can access full-text non-PDF formats that help machines comprehend research papers and generate the output [ 24 ].
AI platforms may not always have access to all standard treatment guidelines. However, in the present study, it was observed that all three AI platforms generally provided appropriate test items regarding the choice of medications, aligning with recommendations from contemporary guidelines and standard textbooks in pharmacology and therapeutics. The prompts used in the study were specifically focused on the pre-clerkship phase of the undergraduate medical curriculum (and at the time of their graduation) and assessed fundamental core concepts, which were also reflected in the AI outputs. Additionally, the recommended first-line antihypertensive drug classes have been established for several decades, and information regarding their pharmacokinetics, ADRs, and indications is well-documented in the literature.
Different paradigms and learning theories have been proposed to support AI in education. These paradigms include AI- directed (learner as recipient), AI-supported (learner as collaborator), and AI-empowered (learner as leader) that are based on Behaviorism, Cognitive-Social constructivism, and Connectivism-Complex adaptive systems, respectively [ 25 ]. AI techniques have potential to stimulate and advance instructional and learning sciences. More recently a three- level model that synthesizes and unifies existing learning theories to model the roles of AIs in promoting learning process has been proposed [ 26 ]. The different components of our study rely upon these paradigms and learning theories as the theoretical underpinning.
Strengths and limitations
To the best of our knowledge, this is the first study evaluating the utility of AI platforms in generating test items related to a discipline in the undergraduate medical curriculum. We have evaluated the AI’s ability to generate outputs related to most types of assessment in the undergraduate medical curriculum. The key lessons learnt for improving the AI-generated test item quality from the present study are outlined in Table 7 . We used a structured framework for assessing the content validity of the test items. However, we have demonstrated using a single case study (hypertension) as a pilot experiment. We chose to evaluate anti-hypertensive drugs as it is a core learning objective and one of the most common disorders relevant to undergraduate medical curricula worldwide. It would be interesting to explore the output from AI platforms for other common (and uncommon/region-specific) disorders, non-/semi-core objectives, and disciplines other than Pharmacology and Therapeutics. An area of interest would be to look at the content validity of the test items generated for different curricula (such as problem-based, integrated, case-based, and competency-based) during different stages of the learning process. Also, we did not attempt to evaluate the generation of flowcharts, algorithms, or figures for generating test items. Another potential area for exploring the utility of AIs in medical education would be repeated procedural practices such as the administration of drugs through different routes by trainee residents [ 27 ]. Several AI tools have been identified for potential application in enhancing classroom instructions and assessment purposes pending validation in prospective studies [ 28 ]. Lastly, we did not administer the AI-generated test items to students and assessed their performance and so could not comment on the validity of test item discrimination and difficulty indices. Additionally, there is a need to confirm the generalizability of the findings to other complex areas in the same discipline as well as in other disciplines that pave way for future studies. The conceptual framework used in the present study for evaluating the AI-generated test items needs to be validated in a larger population. Future studies may also try to evaluate the variations in the AI outputs with repetition of the same queries.
Notwithstanding ongoing discussions and controversies, AI tools are potentially useful adjuncts to optimize instructional methods, test blueprinting, test item generation, and guidance for test standard-setting appropriate to learners’ stage in the medical program. However, experts need to critically review the content validity of AI-generated output. These challenges and caveats are to be addressed before the use of widespread use of AIs in medical education can be advocated.
Data availability
All the data included in this study are provided as Electronic Supplementary Materials.
Tolsgaard MG, Pusic MV, Sebok-Syer SS, Gin B, Svendsen MB, Syer MD, Brydges R, Cuddy MM, Boscardin CK. The fundamentals of Artificial Intelligence in medical education research: AMEE Guide 156. Med Teach. 2023;45(6):565–73.
Article Google Scholar
Sriwastwa A, Ravi P, Emmert A, Chokshi S, Kondor S, Dhal K, Patel P, Chepelev LL, Rybicki FJ, Gupta R. Generative AI for medical 3D printing: a comparison of ChatGPT outputs to reference standard education. 3D Print Med. 2023;9(1):21.
Azer SA, Guerrero APS. The challenges imposed by artificial intelligence: are we ready in medical education? BMC Med Educ. 2023;23(1):680.
Masters K. Ethical use of Artificial Intelligence in Health Professions Education: AMEE Guide 158. Med Teach. 2023;45(6):574–84.
Nagi F, Salih R, Alzubaidi M, Shah H, Alam T, Shah Z, Househ M. Applications of Artificial Intelligence (AI) in Medical Education: a scoping review. Stud Health Technol Inf. 2023;305:648–51.
Google Scholar
Mehta N, Harish V, Bilimoria K, et al. Knowledge and attitudes on artificial intelligence in healthcare: a provincial survey study of medical students. MedEdPublish. 2021;10(1):75.
Mir MM, Mir GM, Raina NT, Mir SM, Mir SM, Miskeen E, Alharthi MH, Alamri MMS. Application of Artificial Intelligence in Medical Education: current scenario and future perspectives. J Adv Med Educ Prof. 2023;11(3):133–40.
Garg T. Artificial Intelligence in Medical Education. Am J Med. 2020;133(2):e68.
Matheny ME, Whicher D, Thadaney IS. Artificial intelligence in health care: a report from the National Academy of Medicine. JAMA. 2020;323(6):509–10.
Sage Poe. Available at: https://poe.com/Assistant (Accessed on. 3rd June 2023).
Claude-Instant: Available at: https://poe.com/Claude-instant (Accessed on 3rd. June 2023).
ChatGPT: Available at: https://poe.com/ChatGPT (Accessed on 3rd. June 2023).
James PA, Oparil S, Carter BL, Cushman WC, Dennison-Himmelfarb C, Handler J, Lackland DT, LeFevre ML, MacKenzie TD, Ogedegbe O, Smith SC Jr, Svetkey LP, Taler SJ, Townsend RR, Wright JT Jr, Narva AS, Ortiz E. 2014 evidence-based guideline for the management of high blood pressure in adults: report from the panel members appointed to the Eighth Joint National Committee (JNC 8). JAMA. 2014;311(5):507–20.
Eschenhagen T. Treatment of hypertension. In: Brunton LL, Knollmann BC, editors. Goodman & Gilman’s the pharmacological basis of therapeutics. 14th ed. New York: McGraw Hill; 2023.
Shabatura J. September. Using Bloom’s taxonomy to write effective learning outcomes. https://tips.uark.edu/using-blooms-taxonomy/ (Accessed on 19th 2023).
Trainor A, Richards JB. Training medical educators to teach: bridging the gap between perception and reality. Isr J Health Policy Res. 2021;10(1):75.
Boscardin C, Gin B, Golde PB, Hauer KE. ChatGPT and generative artificial intelligence for medical education: potential and opportunity. Acad Med. 2023. https://doi.org/10.1097/ACM.0000000000005439 . (Published ahead of print).
Duong MT, Rauschecker AM, Rudie JD, Chen PH, Cook TS, Bryan RN, Mohan S. Artificial intelligence for precision education in radiology. Br J Radiol. 2019;92(1103):20190389.
Karthikeyan S, O’Connor E, Hu W. Barriers and facilitators to writing quality items for medical school assessments - a scoping review. BMC Med Educ. 2019;19(1):123.
Developing teachers and trainers in undergraduate medical education. Advice supplementary to Tomorrow’s Doctors. (2009). https://www.gmc-uk.org/-/media/documents/Developing_teachers_and_trainers_in_undergraduate_medical_education___guidance_0815.pdf_56440721.pdf (Accessed on 19th September 2023).
Cooper A, Rodman A. AI and Medical Education - A 21st-Century Pandora’s Box. N Engl J Med. 2023;389(5):385–7.
Chanda SS, Banerjee DN. Omission and commission errors underlying AI failures. AI Soc. 2022;17:1–24.
Narayanan A, Kapoor S. ‘GPT-4 and Professional Benchmarks: The Wrong Answer to the Wrong Question’. Substack newsletter. AI Snake Oil (blog). https://aisnakeoil.substack.com/p/gpt-4-and-professional-benchmarks (Accessed on 19th September 2023).
Brainard J. November. As scientists face a flood of papers, AI developers aim to help. Science, 21 2023. doi.10.1126/science.adn0669.
Ouyang F, Jiao P. Artificial intelligence in education: the three paradigms. Computers Education: Artif Intell. 2021;2:100020.
Gibson D, Kovanovic V, Ifenthaler D, Dexter S, Feng S. Learning theories for artificial intelligence promoting learning processes. Br J Edu Technol. 2023;54(5):1125–46.
Guerrero DT, Asaad M, Rajesh A, Hassan A, Butler CE. Advancing Surgical Education: the Use of Artificial Intelligence in Surgical Training. Am Surg. 2023;89(1):49–54.
Lee S. AI tools for educators. EIT InnoEnergy Master School Teachers Conference. 2023. https://www.slideshare.net/ignatia/ai-toolkit-for-educators?from_action=save (Accessed on 24th September 2023).
Download references
Author information
Authors and affiliations.
Department of Pharmacology & Therapeutics, College of Medicine & Medical Sciences, Arabian Gulf University, Manama, Kingdom of Bahrain
Kannan Sridharan & Reginald P. Sequeira
You can also search for this author in PubMed Google Scholar
Contributions
RPS– Conceived the idea; KS– Data collection and curation; RPS and KS– Data analysis; RPS and KS– wrote the first draft and were involved in all the revisions.
Corresponding author
Correspondence to Kannan Sridharan .
Ethics declarations
Ethics approval and consent to participate.
Not applicable as neither there was any interaction with humans, nor any personal data was collected in this research study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1
Rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Sridharan, K., Sequeira, R.P. Artificial intelligence and medical education: application in classroom instruction and student assessment using a pharmacology & therapeutics case study. BMC Med Educ 24 , 431 (2024). https://doi.org/10.1186/s12909-024-05365-7
Download citation
Received : 26 September 2023
Accepted : 28 March 2024
Published : 22 April 2024
DOI : https://doi.org/10.1186/s12909-024-05365-7
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Medical education
- Pharmacology
- Therapeutics
BMC Medical Education
ISSN: 1472-6920
- Submission enquiries: [email protected]
- General enquiries: [email protected]
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- NEWS EXPLAINER
- 25 April 2024
- Correction 26 April 2024
Bird flu in US cows: is the milk supply safe?
- Julian Nowogrodzki
You can also search for this author in PubMed Google Scholar

Cows are milked on a farm in New York State. Credit: Angela Weiss/AFP via Getty
Update: On 25 April, the US Department of Agriculture announced that one in five retail milk samples tested contain remnants of the bird flu virus infecting US cattle.
The outbreak of avian influenza in US dairy cattle shows no signs of slowing. Over the past three weeks, the number of states where cows infected with bird flu have been detected has risen from six to eight. A preprint 1 posted on 16 April reported the discovery of the virus in raw milk from infected cows, and US federal authorities said on Wednesday that the virus had been found in lung tissue collected from a seemingly healthy cow.
Also on Wednesday, US officials confirmed at a media briefing that genomic material from the H5N1 strain, which is causing the outbreak, had been detected in milk sold in shops.
Detection of viral particles in milk sold to consumers suggests that avian flu in cows could “be more widespread than initially thought”, says food scientist Diego Diel at Cornell University in Ithaca, New York. “Increased surveillance and testing in dairies should be an important part of control measures going forward.” Nature looks at the implications for human health and the future of the outbreak.
What does it mean that H5N1 is in retail milk?
It’s still unclear how many milk samples the FDA has tested or where the samples were collected. The agency said that it would release more information in the coming days and weeks.
After it leaves the farm and before it hits the shelves, milk is pasteurized to inactivate pathogens. To detect H5N1, the FDA used a test called quantitative polymerase chain reaction (qPCR), which picks up viral RNA. Because it detects fragments of the viral genome, the test cannot distinguish between living virus and the remnants of dead virus, says dairy scientist Nicole Martin at Cornell University.

Bird flu outbreak in mink sparks concern about spread in people
“The detection of viral RNA does not itself pose a health risk to consumers, and we expect to find this residual genetic material if the virus was there in the raw milk and was inactivated by pasteurization,” she says.
The presence of viral material in commercially available milk does have broader implications, however. There are several possible explanations, says virologist Brian Wasik, also at Cornell University. It could be that the outbreak is more pervasive than farmers realized, and that milk from infected animals is entering the commercial supply. Another possibility, he says, is that “asymptomatic cows that we are not testing are shedding virus into milk”. But it’s also possible that both scenarios are true.
US federal rules require milk from infected cows to be discarded, but it’s not yet clear whether cows often start shedding the virus before they look sick or produce abnormal milk. The 16 April preprint, which has not yet been peer reviewed, includes reports that milk from infected cows is thicker and more yellow than typical milk and that infected animals eat less and produce less milk than usual.
Is milk with traces of H5N1 in it a threat to humans?
There is no definitive evidence that pasteurization kills H5N1, but the method kills viruses that multiply in the gut, which are hardier than flu viruses, says Wasik. “Influenza virus is relatively unstable,” he says, “and is very susceptible to heat.” Pasteurization of eggs, which is done at a lower temperature than pasteurization of milk, does kill H5N1.
It’s possible that pasteurization would be less effective at killing relatively high viral concentrations in milk, says Wasik. Finding out whether this is the case requires experimental data. In the absence of a definitive answer, keeping milk from infected cows out of the commercial supply is extremely important.
When Nature asked when to expect more evidence on whether pasteurization kills H5N1, Janell Goodwin, public-affairs specialist at the FDA in Silver Spring, Maryland, said that the agency and the US Department of Agriculture (USDA) “are working closely to collect and evaluate additional data and information specific to” H5N1.
Is milk spreading bird flu among cows?
USDA researchers have tested nasal swabs, tissue and milk samples of cows from affected dairy herds and have found that milk contained the highest viral concentrations. This indicates that the virus could be spreading through milk droplets.

Bird-flu threat disrupts Antarctic penguin studies
If so, milking equipment could be involved. “The teat cups of a milking machine could transfer remnants of H5N1-containing milk from one cow to the teats of the next cow being milked,” says virologist Thijs Kuiken at Erasmus University Medical Centre in Rotterdam, the Netherlands. “Even if they are washed and disinfected, the levels of virus in the milk of infected cattle are so high that one could not exclude the possibility of infectious virus being transferred from cow to cow by this route.” In fact, in some equipment set-ups, workers spray down milking machines with high-pressure hoses to clean them, which would aerosolize any infected milk, says Wasik.
The USDA website concurs that viral spread is “likely through mechanical means”.
Is enough being done to stop the spread?
The FDA announced on Wednesday that cows must test negative for bird flu before they can be moved across state lines. That might help to stem the outbreak, scientists say. Animals in the US dairy industry move around a lot, Wasik says. Calves are moved to be raised into milk cows, cows are moved when they stop producing milk and farmers sell the animals. Such movement is probably “a main driver” of the outbreak, Wasik says.
Diel would like to see surveillance of bulk milk samples at farms. Wastewater testing and environmental sampling could be useful, too, Wasik says, particularly around farms near outbreaks or farms where cows have been moved. He also advocates for a quarantine or observation period of 24 or 48 hours when cattle are moved to a new farm.
Such surveillance measures “could really buy us time, slow down the outbreak”, says Wasik, so researchers and agencies can “get a better handle on it. Because time is what’s of the essence.”
doi: https://doi.org/10.1038/d41586-024-01221-2
Updates & Corrections
Correction 26 April 2024 : An earlier version of this article incorrectly stated the name of the agency that annnounced that one in five milk samples tested positive for avian-flu remnants.
Hu, X. et al. Preprint at bioRxiv https://doi.org/10.1101/2024.04.16.588916 (2024).
Download references
Reprints and permissions
Related Articles

- Agriculture

Humans and their livestock have sheltered in this Saudi Arabian cave for 10,000 years
News 17 APR 24
The complex polyploid genome architecture of sugarcane
Article 27 MAR 24

How science is helping farmers to find a balance between agriculture and solar farms
Spotlight 19 FEB 24

Bird flu virus has been spreading in US cows for months, RNA reveals
News 27 APR 24

WHO redefines airborne transmission: what does that mean for future pandemics?
News 24 APR 24
Monkeypox virus: dangerous strain gains ability to spread through sex, new data suggest
News 23 APR 24
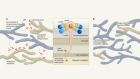
Bacteria deploy umbrella toxins against their competitors
News & Views 17 APR 24

Bird flu outbreak in US cows: why scientists are concerned
News Explainer 08 APR 24
Berlin (DE)
Springer Nature Group
ECUST Seeking Global Talents
Join Us and Create a Bright Future Together!
Shanghai, China
East China University of Science and Technology (ECUST)
Position Recruitment of Guangzhou Medical University
Seeking talents around the world.
Guangzhou, Guangdong, China
Guangzhou Medical University
Junior Group Leader
The Imagine Institute is a leading European research centre dedicated to genetic diseases, with the primary objective to better understand and trea...
Paris, Ile-de-France (FR)
Imagine Institute
Director of the Czech Advanced Technology and Research Institute of Palacký University Olomouc
The Rector of Palacký University Olomouc announces a Call for the Position of Director of the Czech Advanced Technology and Research Institute of P...
Czech Republic (CZ)
Palacký University Olomouc
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
- Skip to main content
- Keyboard shortcuts for audio player
- Your Health
- Treatments & Tests
- Health Inc.
- Public Health
Reproductive rights in America
What's at stake as the supreme court hears idaho case about abortion in emergencies.

Selena Simmons-Duffin

The Supreme Court will hear another case about abortion rights on Wednesday. Protestors gathered outside the court last month when the case before the justices involved abortion pills. Tom Brenner for The Washington Post/Getty Images hide caption
The Supreme Court will hear another case about abortion rights on Wednesday. Protestors gathered outside the court last month when the case before the justices involved abortion pills.
In Idaho, when a pregnant patient has complications, abortion is only legal to prevent the woman's death. But a federal law known as EMTALA requires doctors to provide "stabilizing treatment" to patients in the emergency department.
The Biden administration sees that as a direct conflict, which is why the abortion issue is back – yet again – before the Supreme Court on Wednesday.
The case began just a few weeks after the justices overturned Roe v. Wade in 2022, when the federal Justice Department sued Idaho , arguing that the court should declare that "Idaho's law is invalid" when it comes to emergency abortions because the federal emergency care law preempts the state's abortion ban. So far, a district court agreed with the Biden administration, an appeals court panel agreed with Idaho, and the Supreme Court allowed the strict ban to take effect in January when it agreed to hear the case.

Supreme Court allows Idaho abortion ban to be enacted, first such ruling since Dobbs
The case, known as Moyle v. United States (Mike Moyle is the speaker of the Idaho House), has major implications on everything from what emergency care is available in states with abortion bans to how hospitals operate in Idaho. Here's a summary of what's at stake.
1. Idaho physicians warn patients are being harmed
Under Idaho's abortion law , the medical exception only applies when a doctor judges that "the abortion was necessary to prevent the death of the pregnant woman." (There is also an exception to the Idaho abortion ban in cases of rape or incest, only in the first trimester of the pregnancy, if the person files a police report.)
In a filing with the court , a group of 678 physicians in Idaho described cases in which women facing serious pregnancy complications were either sent home from the hospital or had to be transferred out of state for care. "It's been just a few months now that Idaho's law has been in effect – six patients with medical emergencies have already been transferred out of state for [pregnancy] termination," Dr. Jim Souza, chief physician executive of St. Luke's Health System in Idaho, told reporters on a press call last week.
Those delays and transfers can have consequences. For example, Dr. Emily Corrigan described a patient in court filings whose water broke too early, which put her at risk of infection. After two weeks of being dismissed while trying to get care, the patient went to Corrigan's hospital – by that time, she showed signs of infection and had lost so much blood she needed a transfusion. Corrigan added that without receiving an abortion, the patient could have needed a limb amputation or a hysterectomy – in other words, even if she didn't die, she could have faced life-long consequences to her health.
Attorneys for Idaho defend its abortion law, arguing that "every circumstance described by the administration's declarations involved life-threatening circumstances under which Idaho law would allow an abortion."
Ryan Bangert, senior attorney for the Christian legal powerhouse Alliance Defending Freedom, which is providing pro-bono assistance to the state of Idaho, says that "Idaho law does allow for physicians to make those difficult decisions when it's necessary to perform an abortion to save the life of the mother," without waiting for patients to become sicker and sicker.
Still, Dr. Sara Thomson, an OB-GYN in Boise, says difficult calls in the hospital are not hypothetical or even rare. "In my group, we're seeing this happen about every month or every other month where this state law complicates our care," she says. Four patients have sued the state in a separate case arguing that the narrow medical exception harmed them.
"As far as we know, we haven't had a woman die as a consequence of this law, but that is really on the top of our worry list of things that could happen because we know that if we watch as death is approaching and we don't intervene quickly enough, when we decide finally that we're going to intervene to save her life, it may be too late," she says.
2. Hospitals are closing units and struggling to recruit doctors
Labor and delivery departments are expensive for hospitals to operate. Idaho already had a shortage of providers, including OB-GYNS. Hospital administrators now say the Idaho abortion law has led to an exodus of maternal care providers from the state, which has a population of 2 million people.
Three rural hospitals in Idaho have closed their labor-and-delivery units since the abortion law took effect. "We are seeing the expansion of what's called obstetrical deserts here in Idaho," said Brian Whitlock, president and CEO of the Idaho Hospital Association.
Since Idaho's abortion law took effect, nearly one in four OB-GYNs have left the state or retired, according to a report from the Idaho Physician Well-Being Action Collaborative. The report finds the loss of doctors who specialize in high-risk pregnancies is even more extreme – five of nine full time maternal-fetal medicine specialists have left Idaho.
Administrators say they aren't able to recruit new providers to fill those positions. "Since [the abortion law's] enactment, St. Luke's has had markedly fewer applicants for open physician positions, particularly in obstetrics. And several out-of-state candidates have withdrawn their applications upon learning of the challenges of practicing in Idaho, citing [the law's] enactment and fear of criminal penalties," reads an amicus brief from St. Luke's health system in support of the federal government.
"Prior to the abortion decision, we already ranked 50th in number of physicians per capita – we were already a strained state," says Thomson, the doctor in Boise. She's experienced the loss of OB-GYN colleagues first hand. "I had a partner retire right as the laws were changing and her position has remained open – unfilled now for almost two years – so my own personal group has been short-staffed," she says.
ADF's Bangert says he's skeptical of the assertion that the abortion law is responsible for this exodus of doctors from Idaho. "I would be very surprised if Idaho's abortion law is the sole or singular cause of any physician shortage," he says. "I'm very suspicious of any claims of causality."
3. Justices could weigh in on fetal "personhood"
The state of Idaho's brief argues that EMTALA actually requires hospitals "to protect and care for an 'unborn child,'" an argument echoed in friend-of-the-court briefs from the U.S. Conference of Catholic Bishops and a group of states from Indiana to Wyoming that also have restrictive abortion laws. They argue that abortion can't be seen as a stabilizing treatment if one patient dies as a result.
Thomson is also Catholic, and she says the idea that, in an emergency, she is treating two patients – the fetus and the mother – doesn't account for clinical reality. "Of course, as obstetricians we have a passion for caring for both the mother and the baby, but there are clinical situations where the mom's health or life is in jeopardy, and no matter what we do, the baby is going to be lost," she says.
The Idaho abortion law uses the term "unborn child" as opposed to the words "embryo" or "fetus" – language that implies the fetus has the same rights as other people.

Shots - Health News
The science of ivf: what to know about alabama's 'extrauterine children' ruling.
Mary Ziegler , a legal historian at University of California - Davis, who is writing a book on fetal personhood, describes it as the "North Star" of the anti-abortion rights movement. She says this case will be the first time the Supreme Court justices will be considering a statute that uses that language.
"I think we may get clues about the future of bigger conflicts about fetal personhood," she explains, depending on how the justices respond to this idea. "Not just in the context of this statute or emergency medical scenarios, but in the context of the Constitution."
ADF has dismissed the idea that this case is an attempt to expand fetal rights. "This case is, at root, a question about whether or not the federal government can affect a hostile takeover of the practice of medicine in all 50 states by misinterpreting a long-standing federal statute to contain a hidden nationwide abortion mandate," Bangert says.
4. The election looms large
Ziegler suspects the justices will allow Idaho's abortion law to remain as is. "The Supreme Court has let Idaho's law go into effect, which suggests that the court is not convinced by the Biden administration's arguments, at least at this point," she notes.

Trump backed a federal abortion ban as president. Now, he says he wouldn't sign one
Whatever the decision, it will put abortion squarely back in the national spotlight a few months before the November election. "It's a reminder on the political side of things, that Biden and Trump don't really control the terms of the debate on this very important issue," Zielger observes. "They're going to be things put on everybody's radar by other actors, including the Supreme Court."
The justices will hear arguments in the case on Wednesday morning. A decision is expected by late June or early July.
Correction April 23, 2024
An earlier version of this story did not mention the rape and incest exception to Idaho's abortion ban. A person who reports rape or incest to police can end a pregnancy in Idaho in the first trimester.
- Abortion rights
- Supreme Court
- Marketplace
- Marketplace Morning Report
- Marketplace Tech
- Make Me Smart
- This is Uncomfortable
- The Uncertain Hour
- How We Survive
- Financially Inclined
- Million Bazillion
- Marketplace Minute®
- Corner Office from Marketplace

- Latest Stories
- Collections
- Smart Speaker Skills
- Corrections
- Ethics Policy
- Submissions
- Individuals
- Corporate Sponsorship
- Foundations

Boeing failures are a case study of America’s manufacturing “dark age”

Share Now on:
- https://www.marketplace.org/2024/04/24/boeing-failures-are-a-case-study-of-americas-manufacturing-dark-age/ COPY THE LINK
HTML EMBED:
Sign up for the Marketplace newsletter to get the day’s biggest business stories, our economic analysis, and explainers to help you live smarter, straight to your inbox every weekday evening.
Boeing released its first-quarter financial results Wednesday, and, despite the disastrous Alaska Airlines flight earlier this year and the ensuing public scrutiny the plane manufacturer has faced, the report was not as bad as expected . Revenue came in slightly higher than forecast, but still down 8% from the year before.
Boeing has a storied history that reaches back well over a century. Its current problems raise the question of how we got here — and magnify worries about American manufacturing at large.
Jerry Useem, a contributor to The Atlantic, joined “Marketplace” host Kimberly Adams to talk about Boeing and what he calls the dark age of American manufacturing . An edited transcript of their conversation is below.
Kimberly Adams: You open your piece with a pretty stark comparison of Boeing of the past and Boeing of more recent days. Can you lay that out for me?
Jerry Useem: Well, the comparison is a stark one in that early in Boeing’s existence, it paid very close attention to the process of manufacturing. And the original founder, Bill Boeing, had his office right in the building adjacent to the shop floor and would often stroll over and inspect individual pieces. And what we’ve arrived at now is a situation where the management of Boeing is very much detached from the actual building of its planes.
Adams: Can you briefly sort of take me through the timeline of Boeing, of how they went from this superprestigious company that was doing everything in-house, had all of its engineers very plugged into what was going on, to what we’re learning as these investigations continue about such a disparate chain of events that led to, you know, the door blowing out on the plane and things like that.
Useem: The timeline is kind of a long one. I think it was in the early 2000s that a lot of the key decisions were made. And airplanes are that kind of business where the errors of yesterday take a long time to show up. But around 2005, they really got serious about what they called offloading. And this meant doing less and less of the work in-house. You know, for some of its planes, having the wings built elsewhere, having the entire tail section built elsewhere. But they took this further and further. And I’d say, prior to the accident, it was beginning to recognize that it had taken this too far and was beginning to sort of reintegrate and try to make some steps to bring itself back to kind of its engineering-manufacturing roots.
Adams: How common is what Boeing has done in the rest of American manufacturing?
Useem: I think it’s actually not uncommon. It’s a very visible case for what I think has been a fairly pervasive phenomenon. I mean, as recently as the early 2000s, Intel was seen as the absolute last word in manufacturing prowess, and its CEO frankly admitted that it lost its edge on the shop floor. And they’ve pronounced what he’s called a “death march” to get back to leadership and the actual process of making things better, ensuring quality, etc.
Adams: Yeah, one of the issues you point to in American manufacturing is that power in these companies like Boeing moved from engineers to financial managers. How did that happen, and what’s been the consequence?
Useem: The engineers who first were sort of in charge of the executive suite when the American corporation came to be. Henry Ford and Bill Boeing, these were people who arose from the shop floor. And over time, it’s been pretty well documented that the CEO class came to be populated by people out of the finance function, which meant they spoke the language of numbers and accounting, they didn’t speak the language of engineering. And so those best equipped to understand how to put things together were no longer in charge. And I think that’s a process that’s now gone too long, gone too far.
Adams: What’s the lesson for other American manufacturers to take from this situation? Not just, you know, what Boeing is doing right now, but how Boeing got to this point?
Useem: I think the lesson is ignore the process of making at your peril. You know, a lot of people have set up the Boeing story as sort of a case of a company that’s put costs in front of quality. But the thing is, really what they’ve done is actually made it more expensive for themselves. I mean, they’ve had their production lines shut down. This is extremely costly. So you’ve got to find sustainable ways to keep on driving costs down, keep on improving quality. So it’s a matter of: Are you going to do it bluntly, by getting rid of your experienced workers, your advanced machine tools, and the managerial attention required to make those continual improvements? Or are you going to lean into it?
Stories You Might Like

Truce on Airbus/Boeing subsidies lets sides focus on emerging competitor

Boeing’s 737 Max troubles drag on

Boeing earnings descend, thanks to inflation and lingering supply chain problems

Boeing hits bumpy air as it tries to meet rising demand for aircraft

After preliminary report of the Ethiopian Airlines crash, what’s next for Boeing?

Durable goods slump in April
There’s a lot happening in the world. Through it all, Marketplace is here for you.
You rely on Marketplace to break down the world’s events and tell you how it affects you in a fact-based, approachable way. We rely on your financial support to keep making that possible.
Your donation today powers the independent journalism that you rely on . For just $5/month, you can help sustain Marketplace so we can keep reporting on the things that matter to you.
Also Included in
- Manufacturing
Latest Episodes From Our Shows

The TikTok ban is poised to make the U.S.-China divide even starker

The WIC family food program is getting a refresh, but requirements are still tough to navigate

Why does the world want dollars? Because of high interest rates, thriving economy in U.S.

Recent college grads see rise in unemployment
NTRS - NASA Technical Reports Server
Available downloads, related records.

IMAGES
VIDEO
COMMENTS
Affiliations 1 Laboratory of Virology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, MT 59840, USA; Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Oxford OX3 7BN, UK.; 2 Laboratory of Virology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, MT 59840, USA ...
Case Study: Painful, Purulent Eye of a 56-Year-Old Male. A 56-year-old male presented with 1-day history of pruritic, painful right eye with associated mucopurulent discharge, blurry vision, headache and photosensitivity. ASM is a nonprofit professional society that publishes scientific journals and advances microbiology through advocacy ...
This case study shows an example of the changes that can happen within an immunocompromised patient who is infected with COVID-19 multiple times. Furthermore, this case demonstrates how simultaneous coinfection with two lineages of COVID-19 can lead to unclear lineage assignment by standard methods, which are resolved by further investigation.
Coronavirus disease 2019 (COVID-19) is an acute infection of the respiratory tract that emerged in late 2019 1, 2. Initial outbreaks in China involved 13.8% of cases with severe courses, and 6.1% ...
Mini-case history studies can be used as classroom exercises or given to students to work through as part of a written examination. In this activity, 26 mini-cases divided into two sections, DNA-containing viruses (13 cases) and RNA-containing ... Subsequently, when each new sub-section of virology is covered, use one or two mini-case histories ...
Journal of Medical Virology is a clinical virology journal focused ... A case study of COVID-19. Amirhoshang Hoseinpour Dehkordi ... Spain, Italy, and Germany are 308850, 126168, 124632, and 96092, respectively. Calculating the total case fatality rate (CFR) of Italy (4th April 2020), about 13.3% of confirmed cases have passed away. ...
The study results indicate that IgM was positive with an accuracy of 64.3%, a sensitivity of 44.4% and a specificity of 100% in 28 of 63 samples. The sample identification of 52 cases also showed a positive IgG test with a sensitivity of 82.54%, a specificity of 100% and an accuracy of 88.8%.
As of 25 January 2022, over 349 million individuals have received a confirmed diagnosis of covid-19, with over 5.59 million confirmed deaths associated with the SARS-CoV-2 virus. The covid-19 pandemic has prompted an extensive global effort to study the molecular evolution of the virus and develop vaccines to prevent its spread. Although rigorous determination of SARS-CoV-2 infectivity remains ...
In this case series of five patients with COVID-19, we illustrated three different clinical and biological types of evolution: first, mild cases through two paucisymptomatic patients aged younger than 50 years who were diagnosed early, with high viral load in nasopharyngeal samples, suggesting a significant shedding of SARS-CoV-2, reflected by ...
Virology has largely focused on viruses that are pathogenic to humans or to the other species that we care most about. There is no doubt that this has been a worthwhile investment. But many transformative advances have been made through the in-depth study of relatively obscure viruses that do not appear on lists of prioritized pathogens. In this review, I highlight the benefits that can accrue ...
This case study is written by Rachael Tarlinton, an Associate Professor in Veterinary Virology at the University of Nottingham. Her account of the pandemic expresses the challenges of teaching, supervising and researching throughout lockdowns; how the pressure of this affected many researchers' mental health; and the media engagement duties ...
In this week's case study of Dr. Jude Kong, we learn how modeling helped one community make informed decisions about public health policy during the COVID-19 pandemic and saved lives. Finally, we look at how diseases and pandemics affect different communities differently, and how public health experts are working to address health inequities.
Citation: Virology Journal 2024 21:98 Content type: Review Published on: 26 April 2024. View Full Text ... Data from the COVID-19 clinical control case studies showed tha... Authors: Nozethu Mjokane, Eric O. Akintemi, Saheed Sabiu, Onele M. N. Gcilitshana, Jacobus Albertyn, Carolina H. Pohl ...
Case reports submitted to Virology Journal should make a contribution to medical knowledge and must have educational value or highlight the need for a change in clinical practice or diagnostic/prognostic approaches. ... The Background section should explain the background to the case report or study, its aims, a summary of the existing literature.
Morassi et al., in a case series, reported biochemical evidence of coagulopathy in more than 65% of patients with COVID-19 . Stroke usually developed within a month of onset of the symptoms of COVID-19. In different studies, it was seen that SARS-CoV-2 infection was an independent risk factor for stroke in hospitalized patients [85,86,87,88].
A decade after the first studies were performed it is justifiable to claim that molecular techniques have revolutionised the work of the clinical virology laboratory. Hitherto, the role of the virology laboratory was often a retrospective diagnosis based on virus isolation and serology. Nevertheless, the epidemiological data collected in this way justified the continued activity of clinical ...
Case study - Biobanking in Virology: From Retrospective Sample Collection to Epidemiology, Diagnosis and Research Collection, anonymization and demographic classiication of samples by age, gender, and region can be tricky at a high scale. Cerba Research and Viroclinics-DDL plan to target patients presenting for SARS-CoV-2 testing and run
Virology Case Study - Answer . HISTORY. A 23 year-old male security guard working in a small department store was stabbed with a needle on a syringe of an intravenous drug addict he was arresting for shoplifting. Two months later he developed general malaise, with nausea and vomiting. He lost his appetite and began to lose weight.
Case studies of Virology and Retroviorology. Alain L Fymat An infectiqon might be a submicroscopic infective specialist that reproduces just inside the living cells of a living being. Infections can contaminate a wide range of living things, from creatures and plants to microorganisms, including microscopic organisms and archaea.
Study the viral life cycle or the comorbidities initiated by viral infection when you pursue your PhD in Molecular Virology at Case Western Reserve, one of the top research universities in the U.S. Our interdisciplinary approach to biomedical sciences, through our Biomedical Sciences Training Program, allows you to focus on molecular virology ...
The requirements for a PhD degree in Molecular Virology include course work, participation in formal and informal seminars, a research qualifying examination, and the PhD thesis. In addition to the core curriculum, students are required to complete a minimum of sixteen additional credit hours of advanced course work, including Introduction to ...
case Pic 1. case pic 2. SNAP Parvo test Case Pic 3. Case pic 4 Use PCR if Parvo test is negative. Case pic 5 (Targeting a partial region of VPF2 gene using PCR) Case pic 6 Sequencing of CPV2 genome - reading the viral DNA. Case pic 7. Case pic 8 Identification of CPV 2 Variant. Case pic 9 CPV 2a in Saint Kitts/Nevis.
Study Flashcards On Virology: Case Studies at Cram.com. Quickly memorize the terms, phrases and much more. Cram.com makes it easy to get the grade you want! Virology: Case Studies Flashcards - Cram.com
The term 'revolutionary' has a unique connotation in India's struggle for national liberation. It refers to those freedom fighters who scrupulously believed in the efficacy of armed resistance to overthrow British rule in India and justified employing extremist techniques to achieve the objective.
Artificial intelligence (AI) tools are designed to create or generate content from their trained parameters using an online conversational interface. AI has opened new avenues in redefining the role boundaries of teachers and learners and has the potential to impact the teaching-learning process. In this descriptive proof-of- concept cross-sectional study we have explored the application of ...
Finding out whether this is the case requires experimental data. In the absence of a definitive answer, keeping milk from infected cows out of the commercial supply is extremely important.
The case, known as Moyle v.United States (Mike Moyle is the speaker of the Idaho House), has major implications on everything from what emergency care is available in states with abortion bans to ...
A century ago, U.S manufacturers were run by engineers. Now leadership suites are distant from factory floors, says journalist Jerry Useem.
This work is focused on the role of soil moisture in the prediction of tropical cyclones (TCs) approaching land and after landfall. Soil moisture conditions can impact the circulation and structure of an existing tropical cyclone (TC) when part or all of the circulation is over land. For example, dry land surface conditions may lead to faster dissipation of a TC over land (often associated ...