How to Write a Mini Research Paper Outline
Published 16 October, 2023

A mini research paper outline is a great way to organize your thoughts and get started on an assignment. This blog post is going to walk you through the process of writing a mini-research paper outline. It will not only help you with your own work but also give insight into what professors are looking for from their students.

What is the Outline & Significance of Writing it in a mini-research paper?
An outline is significant for all types of research papers . It serves to arrange your thoughts and your entire work prior to writing a research paper . This kind of paper is aimed at scientific research that will prove you to be a scholar that has technical aptitudes to solve core issues and is all set to convey your ideas using scientific approaches and processes. An outline will be a reminder for you to comprise all the necessary subtleties in it. It is “a frame” of the real research paper that will lead you through the whole procedure but how to write a research paper outline ?
Writing a research paper outline for your mini research paper can give a good direction to the students in writing a research paper. But many students do not have the exact idea about the format of the research paper and that is why they fail to write a good outline during mini research paper submission. The structure of the research paper outline could easily be understood by the students with the help of reliable research paper writers of My Research Topics. All the important steps that are part of a research paper outline could easily be written the Outline of Research Paper by students with the help of these experts.
By preliminary dividing your paper into all its basic parts, you will be far more ordered & will not be concerned that you forgot something. In addition, appear at your outline, you will be calmer as after splitting your work into numerous parts. It will not seem so irresistible & perplexing. You can approach all parts during different days & plan your preparations successively which will assist you to meet even tight time limits!
Get professional research paper writing help from expert writers who can help you in scoring high in college & university. Students across the globe can take guidance in writing research paper outlines, research paper introductions , or even complete research paper writing. So if you are not in a motivation to complete your research paper outline in different subjects like sciences, information technology, Economics, Law and Business studies, etc. Take reliable help in research paper writing from My Research Topics Experts.
Why outline writing is a must for a mini research paper
If you are a student who is used to have research paper writing work on a regular basis, it is not a big deal for you to understand the importance of a research paper outline. Sometimes even professors ask their students to write a mini research paper outline before starting the actual research paper.
The major purpose behind writing a research paper outline is to get an idea about the major points of the topic that you have researched that could be included in the research paper. The majority of the time students forget many significant aspects of the research paper due to a lack of a research paper outline. That is why it is very significant to write a little research paper outline for this purpose.
Mini research paper outlines structure tips
If you are asked by your professors to write a mini research paper outline here are some tips that you must follow for this purpose. My Research Topics Experts have given these tips to the students for their outline of the short research paper.
- Always carry out some research on the topic of your research paperwork before starts writing the outline.
- Make sure to use simple vocabulary and plagiarism-free ideas in your research paper.
- Do not write about the things that are written millions of times already, nobody is interested in reading such research papers.
- Be unique and be innovative along with correct sequences of the arguments in your research paper outline.
It can seem quite difficult to cope with this chore, & in such a case, you can constantly rely on an online writing service. But if you have chosen to write on your own keep reading this piece of writing. To be more capable in the details of the structure look through instances for elementary scholars. The outline for a Literary Essay will also assist you. Anyway, the major parts are as follows:
- Introduction
Seems not that tough, right?! But the fact is that all of the points include a broad range of information for you to arrange in your research outline regarding animals, for example.
The Introduction part is one of the most significant ones. Since it presents the reader with the topic of your paper and it is like a hook that draws the reader’s interest. Here you are supposed to talk about the top necessary components like the thesis statement, the clarification of the topic (some major points, general information), and an explanation of the core terms associated with your learning
The Body part is the amplest one and consists of numerous paragraphs or subparts. Here you bring the opinion to support your report. The research methodology is what follows the introduction segment. It provides insight into the means you carried out the research and must comprise the investigation kind and the questionnaire you have fulfilled. Never forget the aims of the investigation that must be also stated in the introduction.
Make certain to comprise the literature overview. Here mention the creative writing you used as a backup to your hypothesis & theories. This part will demonstrate how you can work the terms, theory, and existing evidence. Your chief theme and the selected literature should be adjacent. Demonstrate how your input develops & distends the active works.
Data and analysis generally go after methods and literature. Here present your results & other variables that you have got in the procedure of the survey. Use tables or graphs if required to be more precise and ordered. Interpret your results. Remember to tell the spectators whether your outcomes bring diversity to the whole topic. Outline the drawbacks of the research & its benefits.
The conclusion part generally does not present the spectators with the new information but gives the cursory look at the whole work by summarizing major points in it. Do not forget to talk about the thesis statement again. Formulate the viewpoint for potential research as well.
Read Also: A Guide to Start Research Process
How to write a mini research paper outline?
Here is the guide to writing the University research paper outline by experts to the students. Those who want to write a perfect research paper outline can follow these points.
- Begin with the topic of research and understand it by multiple dimensions.
- Write down the important points that you noticed from the topic.
- If possible try to sift out the issues and problems that are associated with that topic and how to solve them.
- Also, try to research the reasons which are obstructing these solutions to work on practical grounds.
- Now start writing your research paper outline by giving the abstract or reason why you are writing your research paper.
- Also, discuss the main points that you will raise through your research paper and the way to reach the solutions for these problems.
- Finally, mention the way that you are going to follow to know the reality of these problems and why they exist.
This is how a good research paper outline could be written by the students easily. Students can show this outline to their professors and teachers as well.
As mentioned above a mini research paper talks about the main issues that the writer is going to deal with in his research paper. Apart from that, it also discusses the way and strategies that will be used to reach up to the solution of these problems. Resources that students are going to use in writing a research paper are sometimes also disclosed to the professors.
To cap it all we can say that a mini research paper outline is helpful to the students in keeping all the points in mind while writing a research paper so that any points do not go missing which should be there. Research paper guidance from the experts of My Research Topics also assists the students to write a supreme quality research paper. So students can take the assistance of these experts in their assignments in the form of assistance in research paper writing.
Research paper writing help to The scholars by My Research Topics at a reasonable cost is given round the clock. Those who do not have the idea about writing a good outline for a mini research paper can effortlessly approach for the assistance of experts. The moment you ask for assistance in your assignment of the research paper, a team of professionals from My Research Topics will actively start work upon your academic assignment work.
The research paper writing services are given to the students by an expert at a very cheap cost-effective and budget-friendly price. Every type of student whether he or she belongs to a poor financial background or rich background can have access to this help.
Stuck During Your Dissertation
Our top dissertation writing experts are waiting 24/7 to assist you with your university project,from critical literature reviews to a complete PhD dissertation.

Other Related Guides
- Research Project Questions
- Types of Validity in Research – Explained With Examples
- Schizophrenia Sample Research Paper
- Quantitative Research Methods – Definitive Guide
- Research Paper On Homelessness For College Students
- How to Study for Biology Final Examination
- Textual Analysis in Research / Methods of Analyzing Text
A Guide to Start Research Process – Introduction, Procedure and Tips
Research findings – objectives , importance and techniques.
- Topic Sentences in Research Paper – Meaning, Parts, Importance, Procedure and Techniques

Recent Research Guides for 2023

Get 15% off your first order with My Research Topics
Connect with a professional writer within minutes by placing your first order. No matter the subject, difficulty, academic level or document type, our writers have the skills to complete it.

My Research Topics is provides assistance since 2004 to Research Students Globally. We help PhD, Psyd, MD, Mphil, Undergrad, High school, College, Masters students to compete their research paper & Dissertations. Our Step by step mentorship helps students to understand the research paper making process.
Research Topics & Ideas
- Sociological Research Paper Topics & Ideas For Students 2023
- Nurses Research Paper Topics & Ideas 2023
- Nursing Capstone Project Research Topics & Ideas 2023
- Unique Research Paper Topics & Ideas For Students 2023
- Teaching Research Paper Topics & Ideas 2023
- Literary Research Paper Topics & Ideas 2023
- Nursing Ethics Research Topics & Ideas 2023
Research Guide
Disclaimer: The Reference papers provided by the Myresearchtopics.com serve as model and sample papers for students and are not to be submitted as it is. These papers are intended to be used for reference and research purposes only.
Subscribe to the PwC Newsletter
Join the community, edit social preview.
Add a new code entry for this paper
Remove a code repository from this paper.

Mark the official implementation from paper authors
Add a new evaluation result row.
- IMAGE COMPREHENSION
- VISUAL DIALOG
- VISUAL QUESTION ANSWERING
Remove a task

Add a method
- ABSOLUTE POSITION ENCODINGS
- DENSE CONNECTIONS
- LABEL SMOOTHING
- LAYER NORMALIZATION
- LINEAR LAYER
- MULTI-HEAD ATTENTION
- POSITION-WISE FEED-FORWARD LAYER
- RESIDUAL CONNECTION
- SCALED DOT-PRODUCT ATTENTION
- TRANSFORMER
Remove a method
- ABSOLUTE POSITION ENCODINGS -
- DENSE CONNECTIONS -
- LABEL SMOOTHING -
- LAYER NORMALIZATION -
- LINEAR LAYER -
- MULTI-HEAD ATTENTION -
- POSITION-WISE FEED-FORWARD LAYER -
- RESIDUAL CONNECTION -
- SCALED DOT-PRODUCT ATTENTION -
- TRANSFORMER -
Include the markdown at the top of your GitHub README.md file to showcase the performance of the model.
Badges are live and will be dynamically updated with the latest ranking of this paper.
Edit Datasets
Mini-gemini: mining the potential of multi-modality vision language models.
27 Mar 2024 · Yanwei Li , Yuechen Zhang , Chengyao Wang , Zhisheng Zhong , Yixin Chen , Ruihang Chu , Shaoteng Liu , Jiaya Jia · Edit social preview
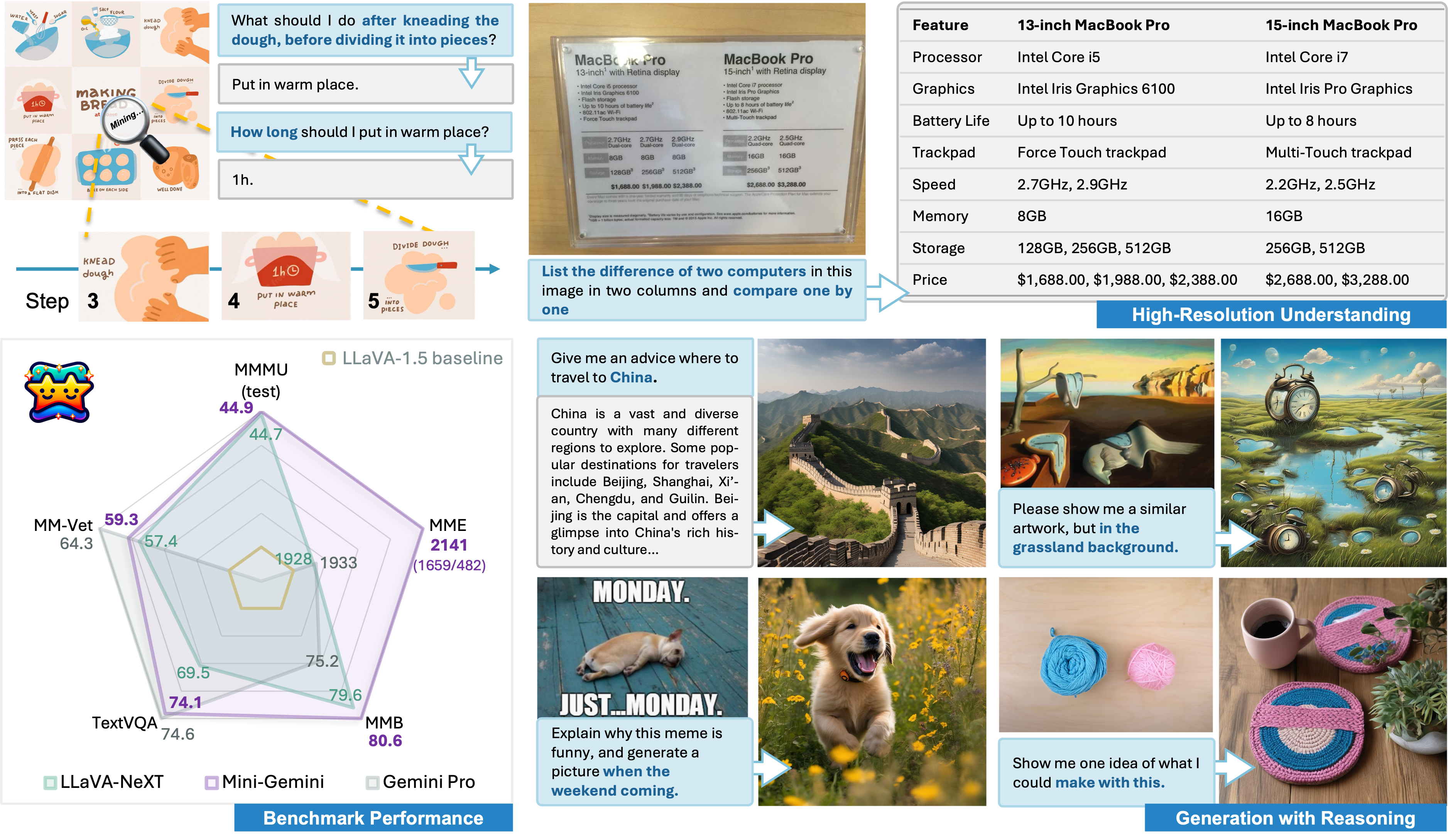
In this work, we introduce Mini-Gemini, a simple and effective framework enhancing multi-modality Vision Language Models (VLMs). Despite the advancements in VLMs facilitating basic visual dialog and reasoning, a performance gap persists compared to advanced models like GPT-4 and Gemini. We try to narrow the gap by mining the potential of VLMs for better performance and any-to-any workflow from three aspects, i.e., high-resolution visual tokens, high-quality data, and VLM-guided generation. To enhance visual tokens, we propose to utilize an additional visual encoder for high-resolution refinement without increasing the visual token count. We further construct a high-quality dataset that promotes precise image comprehension and reasoning-based generation, expanding the operational scope of current VLMs. In general, Mini-Gemini further mines the potential of VLMs and empowers current frameworks with image understanding, reasoning, and generation simultaneously. Mini-Gemini supports a series of dense and MoE Large Language Models (LLMs) from 2B to 34B. It is demonstrated to achieve leading performance in several zero-shot benchmarks and even surpasses the developed private models. Code and models are available at https://github.com/dvlab-research/MiniGemini.
Code Edit Add Remove Mark official
Tasks edit add remove, datasets edit.

Results from the Paper Edit

Methods Edit Add Remove
- Create Account
Main navigation dropdown
Publications, miniseries: ai for iot, publication date, manuscript submission deadline, call for ai for iot articles.
The IEEE Internet of Things Magazine (IEEE IoTM) is soliciting articles for its mini-series on “AI for IoT”. The proliferation of IoT devices and sensors, coupled with advancements in AI algorithms and computing technologies, has paved the way for a new era of intelligent IoT systems. AI techniques are increasingly integrated into IoT architectures to enable advanced analytics, autonomous decision-making, and adaptive behaviors. From smart homes and cities to industrial automation and healthcare, AI-powered IoT solutions are revolutionizing the way we interact with and leverage data from connected devices, driving innovation, efficiency, and sustainability. This mini-series aims to explore the intersection of AI and IoT, covering cutting-edge research, real-world applications, and best practices in leveraging AI to enhance IoT systems and services.
Articles for this mini-series are invited to cover a wide range of topics, including but not limited to:
- AI-enabled IoT Applications and Use Cases : Explore innovative applications and use cases where AI enhances IoT functionalities and capabilities, spanning smart healthcare, intelligent transportation, precision agriculture, industrial automation, environmental monitoring, and more.
- AI-driven Data Analytics and Decision-making: Investigate AI techniques for processing, analyzing, and deriving actionable insights from IoT-generated data streams, enabling predictive maintenance, anomaly detection, personalized recommendations, and intelligent automation.
- Generative AI and Large Language Models (LLMs) for IoT : Study the applications of generative AI in IoT environments; explore how LLMs, such as generative pre-trained transformer (GPT) models, can be utilized to generate synthetic data, enhance natural language understanding, and support human-machine interaction in IoT systems.
- Edge AI and Distributed Intelligence: Discuss the integration of AI algorithms and models at the edge of IoT networks, enabling real-time inference, adaptive learning, and autonomous decision-making closer to data sources, and minimizing latency and bandwidth requirements.
- AI-empowered Robotics and Sensing for IoT : Explore the integration of AI with robotics and sensing technologies in IoT systems. Introduce advancements in sensor technologies and data fusion techniques that enable intelligent data collection, processing, and analysis in dynamic environments.
- Privacy, Security, and Trustworthiness : Address the privacy and security implications of AI-enabled IoT systems, including data privacy, confidentiality, integrity, and authenticity. Investigate trustworthiness in AI-enabled IoT systems, emphasizing the need for reliability, transparency, explainability, accountability, and fairness.
- Standardization and Interoperability : Discuss the challenges and opportunities in standardizing AI-enabled IoT technologies to ensure interoperability, compatibility, and seamless integration across heterogeneous IoT ecosystems. Explore emerging standards, protocols, and frameworks for facilitating collaboration and interoperability among AI and IoT technologies.
- Demonstrations, Proof-of-Concepts, and Deployments : Present innovative demonstrations and proof-of-concepts showcasing the integration of AI technologies with IoT systems. Share insights and best practices for deploying AI-powered IoT solutions in diverse environments, including smart cities, healthcare, agriculture, manufacturing, transportation, and energy.
- Regulation and Policy : Explore the regulatory and policy landscape surrounding AI and IoT technologies, including data governance, consumer protection, liability, accountability, and ethical considerations. Discuss the role of regulatory bodies, industry consortia, and international organizations in shaping ethical, legal, and policy frameworks to ensure the responsible development, deployment, and use of AI-powered IoT systems.
Authors should keep in mind that the intended audience consists of all the members of the IoT community. Hence, articles must be understandable by the general IoT practitioner/researcher, independent of technical or business specialty and are expected to add to the knowledge base or best practices of the IoT community. Mathematical material should be avoided; instead, references to papers containing the relevant mathematics should be provided when applicable.
In addition to the above, the IEEE IoTM general paper submission guidelines and author guidelines for manuscript development and submission over manuscript central must be carefully abided by.
Note: IoTM does not have a specific template and does not require manuscripts to be submitted in any specific layout. However, authors can use the template for IEEE Transactions to get a rough estimate of the page count.
Navigation Menu
Search code, repositories, users, issues, pull requests..., provide feedback.
We read every piece of feedback, and take your input very seriously.
Saved searches
Use saved searches to filter your results more quickly.
To see all available qualifiers, see our documentation .
- Notifications
Official repo for "Mini-Gemini: Mining the Potential of Multi-modality Vision Language Models"
dvlab-research/MGM
Folders and files, repository files navigation.
The framework supports a series of dense and MoE Large Language Models (LLMs) from 2B to 34B with image understanding, reasoning, and generation simultaneously. We build this repo based on LLaVA.
- [04/15] 🔥 The Hugging Face demo is available. It's a 13B-HD version, welcome to watch and try.
- [03/28] 🔥 Mini-Gemini is coming! We release the paper , demo , code , models , and data !
Preparation
Acknowledgement.
We provide some selected examples in this section. More examples can be found in our project page . Feel free to try our online demo !

Please follow the instructions below to install the required packages.
NOTE: If you want to use the 2B version, please ensure to install the latest version Transformers (>=4.38.0).
- Clone this repository
- Install Package
- Install additional packages for training cases
The framework is conceptually simple: dual vision encoders are utilized to provide low-resolution visual embedding and high-resolution candidates; patch info mining is proposed to conduct patch-level mining between high-resolution regions and low-resolution visual queries; LLM is utilized to marry text with images for both comprehension and generation at the same time.

We provide all our fully finetuned models on Stage 1 and 2 data:
Here are the pretrained weights on Stage 1 data only:
We provide the processed data for the model training. For model pretraining, please download the following the training image-based data and organize them as:
-> means put the data in the local folder.
- LLaVA Images -> data/MGM-Pretrain/images , data/MGM-Finetune/llava/LLaVA-Pretrain/images
- ALLaVA Caption -> data/MGM-Pretrain/ALLaVA-4V
For model finetuning, please download the following the instruction data and organize them as:
- COCO train2017 -> data/MGM-Finetune/coco
- GQA -> data/MGM-Finetune/gqa
- OCR-VQA ( we save all files as .jpg ) -> data/MGM-Finetune/ocr_vqa
- TextVQA (not included for training) -> data/MGM-Finetune/textvqa
- VisualGenome part1 , VisualGenome part2 -> data/MGM-Finetune/vg
- ShareGPT4V-100K -> data/MGM-Finetune/sam , share_textvqa , wikiart , web-celebrity , web-landmark
- LAION GPT4V -> data/MGM-Finetune/gpt4v-dataset
- ALLaVA Instruction -> data/MGM-Pretrain/ALLaVA-4V
- DocVQA -> data/MGM-Finetune/docvqa
- ChartQA -> data/MGM-Finetune/chartqa
- DVQA -> data/MGM-Finetune/dvqa
- AI2D -> data/MGM-Finetune/ai2d
For model evaluation, please follow this link for preparation. We use some extra benchmarks for evaluation. please download the following the training image-based data and organize them as:
- MMMU -> data/MGM-Eval/MMMU
- MMB -> data/MGM-Eval/MMB
- MathVista -> data/MGM-Eval/MathVista
Please put the pretrained data, finetuned data, and eval data in MGM-Pretrain , MGM-Finetune , and MGM-Eval subset following Structure .
For meta info, please download the following files and organize them as in Structure .
IMPORTANT: mgm_generation_pure_text.json is a generation-related subset. DO NOT merge it with mgm_instruction.json as it is already included in it. You may merge this file with your customized LLM/VLM SFT dataset to enable the reasoning generation ability.
Pretrained Weights
We recommend users to download the pretrained weights from the following link CLIP-Vit-L-336 , OpenCLIP-ConvNeXt-L , Gemma-2b-it , Vicuna-7b-v1.5 , Vicuna-13b-v1.5 , Mixtral-8x7B-Instruct-v0.1 , and Nous-Hermes-2-Yi-34B , and put them in model_zoo following Structure .
The folder structure should be organized as follows before training.
The training process consists of two stages: (1) feature alignment stage: bridge the vision and language tokens; (2) instruction tuning stage: teach the model to follow multimodal instructions.
Our models are trained on 8 A100 GPUs with 80GB memory. To train on fewer GPUs, you can reduce the per_device_train_batch_size and increase the gradient_accumulation_steps accordingly. Always keep the global batch size the same: per_device_train_batch_size x gradient_accumulation_steps x num_gpus .
Please make sure you download and organize the data following Preparation before training.
NOTE: Please set hostfile for 2 machine training and hostfile_4 for 4 machine training.
If you want to train and finetune the framework, please run the following command for MGM-7B with image size 336:
or for MGM-13B with image size 336:
Because we reuse the pre-trained projecter weights from the MGM-7B, you can directly use the MGM-7B-HD with image size 672 for stage-2 instruction tuning:
Please find more training scripts of gemma , llama , mixtral , and yi in scripts/ .
We perform evaluation on several image-based benchmarks. Please download the evaluation data following Preparation and organize them as in Structure .
If you want to evaluate the model on image-based benchmarks, please use the scripts in scripts/MODEL_PATH/eval . For example, run the following command for TextVQA evaluation with MGM-7B-HD:
Please find more evaluation scripts in scripts/MODEL_PATH .
CLI Inference
Chat with images without the need of Gradio interface. It also supports multiple GPUs, 4-bit and 8-bit quantized inference. With 4-bit quantization. Please make sure you have installed diffusers and PaddleOCR (only for better experience with OCR), and try this for image and generation inference:
or try this better experience with OCR (make sure you have installed PaddleOCR ):
or try this for inference with generation (make sure you have installed diffusers ):
You can also try 8bit or even 4bit for efficient inference
Gradio Web UI
Here, we adopt the Gradio UI similar to that in LLaVA to provide a user-friendly interface for our models. To launch a Gradio demo locally, please run the following commands one by one. If you plan to launch multiple model workers to compare between different checkpoints, you only need to launch the controller and the web server ONCE .
Launch a controller
Launch a gradio web server..
You just launched the Gradio web interface. Now, you can open the web interface with the URL printed on the screen. You may notice that there is no model in the model list. Do not worry, as we have not launched any model worker yet. It will be automatically updated when you launch a model worker.
Launch a model worker
This is the actual worker that performs the inference on the GPU. Each worker is responsible for a single model specified in --model-path .
Wait until the process finishes loading the model and you see "Uvicorn running on ...". Now, refresh your Gradio web UI, and you will see the model you just launched in the model list.
You can launch as many workers as you want, and compare between different models in the same Gradio interface. Please keep the --controller the same, and modify the --port and --worker to a different port number for each worker.
If you are using an Apple device with an M1 or M2 chip, you can specify the mps device by using the --device flag: --device mps .
Launch a model worker (Multiple GPUs, when GPU VRAM <= 24GB)
If the VRAM of your GPU is less than 24GB (e.g., RTX 3090, RTX 4090, etc.), you may try running it with multiple GPUs. Our latest code base will automatically try to use multiple GPUs if you have more than one GPU. You can specify which GPUs to use with CUDA_VISIBLE_DEVICES . Below is an example of running with the first two GPUs.
Launch a model worker (4-bit, 8-bit inference, quantized)
You can launch the model worker with quantized bits (4-bit, 8-bit), which allows you to run the inference with reduced GPU memory footprint. Note that inference with quantized bits may not be as accurate as the full-precision model. Simply append --load-4bit or --load-8bit to the model worker command that you are executing. Below is an example of running with 4-bit quantization.
We provide some examples in this section. More examples can be found in our project page .
Hi-Resolution Understanding

Generation with Reasoning

If you find this repo useful for your research, please consider citing the paper
This project is not affiliated with Google LLC.
We would like to thank the following repos for their great work:
- This work is built upon the LLaVA .
- This work utilizes LLMs from Gemma , Vicuna , Mixtral , and Nous-Hermes .
The data and checkpoint is intended and licensed for research use only. They are also restricted to uses that follow the license agreement of LLaVA, LLaMA, Vicuna and GPT-4. The dataset is CC BY NC 4.0 (allowing only non-commercial use) and models trained using the dataset should not be used outside of research purposes.
Contributors 5
- Python 87.0%
- JavaScript 1.8%

Mini review
The Mini review article type denotes a review with a more concise format compared with a standard review article.
Preparing your manuscript
The title page should:
- present a title that includes, if appropriate, the research design or for non-research studies: a description of what the article reports
- if a collaboration group should be listed as an author, please list the group name as an author and include the names of the individual members of the group in the “Acknowledgements” section in accordance with the instructions below
- indicate the corresponding author
The abstract should briefly summarize the aim, findings or purpose of the article. Please minimize the use of abbreviations and do not cite references in the abstract.
Beginning January 2022, we welcome submissions that include a graphical abstract (GA). This image should be a summary of the findings from the presented research allowing readers to quickly deduce the content in a visual format. The purpose is to highlight your work – draw the readers in quickly so they want to read more.
This graphical abstract figure (drawing, structure, or reaction scheme), preferably in color (free), will be used in the Table of Contents and in the abstract section on the title page of the article. Cover art is often chosen from graphical abstract figures.
For the GA, include a short title and description (about 50 words).The figure should be in one of the following file types: .tiff, .eps, .jpg, .bmp, .doc, or .pdf. It should be 8 cm (3.15 inches) wide x 4 cm (1.57 inches) high when printed at full scale (100%), and should have high quality image and text. Please insure that the illustration maintains this aspect ratio and is still informative upon reduction.
Please supply the GA figure at 100% using the following specifications/sizes:
- 300 dpi – halftone
- 600 dpi - with text
- 600 dpi - combine halftone and text (embedded text)
- 1200 dpi - bitmap (pure text and lines (b/w))
- 300/600/1200 dpi - combine embedded images and vector objects
- For "rastered" images (.pdf, .doc, .bmp, .jpg), the resolution should be at least 300 dpi.
Keywords
Three to ten keywords representing the main content of the article.
This should contain the body of the article, and may also be broken into subsections with short, informative headings.
List of abbreviations
If abbreviations are used in the text they should be defined in the text at first use, and a list of abbreviations should be provided.
Declarations
All manuscripts must contain the following sections under the heading 'Declarations':
Availability of data and materials
Competing interests, authors' contributions, acknowledgements.
- Authors' information (optional)
Please see below for details on the information to be included in these sections.
If any of the sections are not relevant to your manuscript, please include the heading and write 'Not applicable' for that section.
All manuscripts must include an ‘Availability of data and materials’ statement. Data availability statements should include information on where data supporting the results reported in the article can be found including, where applicable, hyperlinks to publicly archived datasets analysed or generated during the study. By data we mean the minimal dataset that would be necessary to interpret, replicate and build upon the findings reported in the article. We recognise it is not always possible to share research data publicly, for instance when individual privacy could be compromised, and in such instances data availability should still be stated in the manuscript along with any conditions for access.
Data availability statements can take one of the following forms (or a combination of more than one if required for multiple datasets):
- The datasets generated and/or analysed during the current study are available in the [NAME] repository, [PERSISTENT WEB LINK TO DATASETS]
- The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
- All data generated or analysed during this study are included in this published article [and its supplementary information files].
- The datasets generated and/or analysed during the current study are not publicly available due [REASON WHY DATA ARE NOT PUBLIC] but are available from the corresponding author on reasonable request.
- Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.
- The data that support the findings of this study are available from [third party name] but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of [third party name].
- Not applicable. If your manuscript does not contain any data, please state 'Not applicable' in this section.
More examples of template data availability statements, which include examples of openly available and restricted access datasets, are available here .
SpringerOpen also requires that authors cite any publicly available data on which the conclusions of the paper rely in the manuscript. Data citations should include a persistent identifier (such as a DOI) and should ideally be included in the reference list. Citations of datasets, when they appear in the reference list, should include the minimum information recommended by DataCite and follow journal style. Dataset identifiers including DOIs should be expressed as full URLs. For example:
Hao Z, AghaKouchak A, Nakhjiri N, Farahmand A. Global integrated drought monitoring and prediction system (GIDMaPS) data sets. figshare. 2014. http://dx.doi.org/10.6084/m9.figshare.853801
With the corresponding text in the Availability of data and materials statement:
The datasets generated during and/or analysed during the current study are available in the [NAME] repository, [PERSISTENT WEB LINK TO DATASETS]. [Reference number]
If you wish to co-submit a data note describing your data to be published in BMC Research Notes , you can do so by visiting our submission portal . Data notes support open data and help authors to comply with funder policies on data sharing. Co-published data notes will be linked to the research article the data support ( example ).
All financial and non-financial competing interests must be declared in this section.
See our editorial policies for a full explanation of competing interests. If you are unsure whether you or any of your co-authors have a competing interest please contact the editorial office.
Please use the authors’ initials to refer to each authors' competing interests in this section.
If you do not have any competing interests, please state "The authors declare that they have no competing interests" in this section.
All sources of funding for the research reported should be declared. If the funder has a specific role in the conceptualization, design, data collection, analysis, decision to publish, or preparation of the manuscript, this should be declared.
The individual contributions of authors to the manuscript should be specified in this section. Guidance and criteria for authorship can be found in our editorial policies .
Please use initials to refer to each author's contribution in this section, for example: "FC analyzed and interpreted the patient data regarding the hematological disease and the transplant. RH performed the histological examination of the kidney, and was a major contributor in writing the manuscript. All authors read and approved the final manuscript."
Please acknowledge anyone who contributed towards the article who does not meet the criteria for authorship including anyone who provided professional writing services or materials.
Authors should obtain permission to acknowledge from all those mentioned in the Acknowledgements section.
See our editorial policies for a full explanation of acknowledgements and authorship criteria.
If you do not have anyone to acknowledge, please write "Not applicable" in this section.
Group authorship (for manuscripts involving a collaboration group): if you would like the names of the individual members of a collaboration Group to be searchable through their individual PubMed records, please ensure that the title of the collaboration Group is included on the title page and in the submission system and also include collaborating author names as the last paragraph of the “Acknowledgements” section. Please add authors in the format First Name, Middle initial(s) (optional), Last Name. You can add institution or country information for each author if you wish, but this should be consistent across all authors.
Authors' information
This section is optional.
You may choose to use this section to include any relevant information about the author(s) that may aid the reader's interpretation of the article, and understand the standpoint of the author(s). This may include details about the authors' qualifications, current positions they hold at institutions or societies, or any other relevant background information. Please refer to authors using their initials. Note this section should not be used to describe any competing interests.
Footnotes should be designated within the text using a superscript number. It is not allowed to use footnotes for references/citations.
Examples of the Basic Springer reference style are shown below.
See our editorial policies for author guidance on good citation practice.
Web links and URLs: All web links and URLs, including links to the authors' own websites, should be given a reference number and included in the reference list rather than within the text of the manuscript. They should be provided in full, including both the title of the site and the URL, as well as the date the site was accessed, in the following format: The Mouse Tumor Biology Database. http://tumor.informatics.jax.org/mtbwi/index.do . Accessed 20 May 2013. If an author or group of authors can clearly be associated with a web link, such as for weblogs, then they should be included in the reference.
Example reference style:
Article within a journal
Smith J, Jones M Jr, Houghton L (1999) Future of health insurance. N Engl J Med 965:325-329.
Article by DOI (with page numbers)
Slifka MK, Whitton JL (2000) Clinical implications of dysregulated cytokine production. J Mol Med 78:74-80. doi:10.1007/s001090000086.
Article by DOI (before issue publication and with page numbers)
Slifka MK, Whitton JL (2000) Clinical implications of dysregulated cytokine production. J Mol Med. doi:10.1007/s001090000086.
Article in electronic journal by DOI (no paginated version)
Slifka MK, Whitton JL (2000) Clinical implications of dysregulated cytokine production. Dig J Mol Med. doi:10.1007/s801090000086.
Journal issue with issue editor
Smith J (ed) (1998) Rodent genes. Mod Genomics J 14(6):126-233.
Journal issue with no issue editor
Mod Genomics J (1998) Rodent genes. Mod Genomics J 14(6):126-233.
Book chapter, or an article within a book
Brown B, Aaron M (2001) The politics of nature. In: Smith J (ed) The rise of modern genomics, 3rd edn. Wiley, New York.
Complete book, authored
South J, Blass B (2001) The future of modern genomics. Blackwell, London.
Complete book, edited
Smith J, Brown B (eds) (2001) The demise of modern genomics. Blackwell, London.
Complete book, also showing a translated edition [Either edition may be listed first.]
Adorno TW (1966) Negative Dialektik. Suhrkamp, Frankfurt. English edition: Adorno TW (1973) Negative Dialectics (trans: Ashton EB). Routledge, London.
Chapter in a book in a series without volume titles
Schmidt H (1989) Testing results. In: Hutzinger O (ed) Handbook of environmental chemistry, vol 2E. Springer, Heidelberg, p 111.
Chapter in a book in a series with volume titles
Smith SE (1976) Neuromuscular blocking drugs in man. In: Zaimis E (ed) Neuromuscular junction. Handbook of experimental pharmacology, vol 42. Springer, Heidelberg, pp 593-660.
OnlineFirst chapter in a series (without a volume designation but with a DOI)
Saito, Yukio, and Hyuga, Hiroyuki. (2007) Rate equation approaches to amplification of enantiomeric excess and chiral symmetry breaking. Topics in Current Chemistry. doi:10.1007/128_2006_108.
Proceedings as a book (in a series and subseries)
Zowghi D (1996) A framework for reasoning about requirements in evolution. In: Foo N, Goebel R (eds) PRICAI'96: topics in artificial intelligence. 4th Pacific Rim conference on artificial intelligence, Cairns, August 1996. Lecture notes in computer science (Lecture notes in artificial intelligence), vol 1114. Springer, Heidelberg, p 157.
Article within conference proceedings with an editor (without a publisher)
Aaron M (1999) The future of genomics. In: Williams H (ed) Proceedings of the genomic researchers, Boston, 1999.
Article within conference proceedings without an editor (without a publisher)
Chung S-T, Morris RL (1978) Isolation and characterization of plasmid deoxyribonucleic acid from Streptomyces fradiae. In: Abstracts of the 3rd international symposium on the genetics of industrial microorganisms, University of Wisconsin, Madison, 4-9 June 1978.
Article presented at a conference
Chung S-T, Morris RL (1978) Isolation and characterization of plasmid deoxyribonucleic acid from Streptomyces fradiae. Paper presented at the 3rd international symposium on the genetics of industrial microorganisms, University of Wisconsin, Madison, 4-9 June 1978.
Norman LO (1998) Lightning rods. US Patent 4,379,752, 9 Sept 1998.
Dissertation
Trent JW (1975) Experimental acute renal failure. Dissertation, University of California.
Book with institutional author
International Anatomical Nomenclature Committee (1966) Nomina anatomica. Excerpta Medica, Amsterdam.
In press article
Major M (2007) Recent developments. In: Jones W (ed) Surgery today. Springer, Dordrecht (in press).
Online document
Doe J (1999) Title of subordinate document. In: The dictionary of substances and their effects. Royal Society of Chemistry. Available via DIALOG. http://www.rsc.org/dose/title of subordinate document. Accessed 15 Jan 1999.
Online database
Healthwise Knowledgebase (1998) US Pharmacopeia, Rockville. http://www.healthwise.org. Accessed 21 Sept 1998.
Supplementary material/private homepage
Doe J (2000) Title of supplementary material. http://www.privatehomepage.com. Accessed 22 Feb 2000.
University site
Doe J (1999) Title of preprint. http://www.uni-heidelberg.de/mydata.html. Accessed 25 Dec 1999.
Doe J (1999) Trivial HTTP, RFC2169. ftp://ftp.isi.edu/in-notes/rfc2169.txt. Accessed 12 Nov 1999.
Organization site
ISSN International Centre (2006) The ISSN register. http://www.issn.org. Accessed 20 Feb 2007.
General formatting information
Manuscripts must be written in concise English. For help on scientific writing, or preparing your manuscript in English, please see Springer's Author Academy .
Quick points:
- Use double line spacing
- Include line and page numbering
- Use SI units: Please ensure that all special characters used are embedded in the text, otherwise they will be lost during conversion to PDF
- Do not use page breaks in your manuscript
File formats
The following word processor file formats are acceptable for the main manuscript document:
- Microsoft word (DOC, DOCX)
- Rich text format (RTF)
- TeX/LaTeX
Please note: editable files are required for processing in production. If your manuscript contains any non-editable files (such as PDFs) you will be required to re-submit an editable file if your manuscript is accepted.
For more information, see ' Preparing figures ' below.
Additional information for TeX/LaTeX users
You are encouraged to use the Springer Nature LaTeX template when preparing a submission. A PDF of your manuscript files will be compiled during submission using pdfLaTeX and TexLive 2021. All relevant editable source files must be uploaded during the submission process. Failing to submit these source files will cause unnecessary delays in the production process.
Style and language
For editors and reviewers to accurately assess the work presented in your manuscript you need to ensure the English language is of sufficient quality to be understood. If you need help with writing in English you should consider:
- Getting a fast, free online grammar check .
- Visiting the English language tutorial which covers the common mistakes when writing in English.
- Asking a colleague who is proficient in English to review your manuscript for clarity.
- Using a professional language editing service where editors will improve the English to ensure that your meaning is clear and identify problems that require your review. Two such services are provided by our affiliates Nature Research Editing Service and American Journal Experts . SpringerOpen authors are entitled to a 10% discount on their first submission to either of these services. To claim 10% off English editing from Nature Research Editing Service, click here . To claim 10% off American Journal Experts, click here .
Please note that the use of a language editing service is not a requirement for publication in AAPS Open and does not imply or guarantee that the article will be selected for peer review or accepted. 为便于编辑和评审专家准确评估您稿件中陈述的研究工作,您需要确保文稿英语语言质量足以令人理解。如果您需要英文写作方面的帮助,您可以考虑:
- 获取快速、免费的在线 语法检查 。
- 查看一些有关英语写作中常见语言错误的 教程 。
- 请一位以英语为母语的同事审阅您的稿件是否表意清晰。
- 使用专业语言编辑服务,编辑人员会对英语进行润色,以确保您的意思表达清晰,并提出需要您复核的问题。例如我们的附属机构 Nature Research Editing Service 以及合作伙伴 American Journal Experts 都可以提供此类专业服务。SpringerOpen作者享受首次订单10%优惠,该优惠同时适用于两家公司。您只需点击以下链接即可开始。使用 Nature Research Editing Service的编辑润色10%的优惠服务,请点击 这里 。使用 American Journal Experts的10%优惠服务,请点击 这里 。
请注意,使用语言编辑服务并非在期刊上发表文章的必要条件,这也并不意味或保证文章将被选中进行同行评议或被接受。 エディターと査読者があなたの論文を正しく評価するには、使用されている英語の質が十分であることが必要とされます。英語での論文執筆に際してサポートが必要な場合には、次のオプションがあります:
- 高速なオンライン 文法チェック を無料で受ける。
- 英語で執筆する際のよくある間違いに関する 英語のチュートリアル を参照する。
- 英語を母国語とする同僚に、原稿内の英語が明確であるかをチェックしてもらう。
- プロの英文校正サービスを利用する。校正者が原稿の意味を明確にしたり、問題点を指摘し、英語を向上させます。 Nature Research Editing Service と American Journal Experts の2つは弊社と提携しているサービスです。SpringerOpenのジャーナルの著者は、いずれかのサービスを初めて利用する際に、10%の割引を受けることができます。Nature Research Editing Serviceの10%割引を受けるには、 こちらをクリックしてください 。. American Journal Expertsの10%割引を受けるには、 こちらをクリックしてください 。
英文校正サービスの利用は、このジャーナルに掲載されるための条件ではないこと、また論文審査や受理を保証するものではないことに留意してください。 영어 원고의 경우, 에디터 및 리뷰어들이 귀하의 원고에 실린 결과물을 정확하게 평가할 수 있도록, 그들이 충분히 이해할 수 있을 만한 수준으로 작성되어야 합니다. 만약 영작문과 관련하여 도움을 받기를 원하신다면 다음의 사항들을 고려하여 주십시오:
- 영어 튜토리얼 페이지 에 방문하여 영어로 글을 쓸 때 자주하는 실수들을 확인합니다.
- 귀하의 원고의 표현을 명확히 해줄 영어 원어민 동료를 찾아서 리뷰를 의뢰합니다
- 리뷰에 대비하여, 원고의 의미를 명확하게 해주고 리뷰에서 요구하는 문제점들을 식별해서 영문 수준을 향상시켜주는 전문 영문 교정 서비스를 이용합니다. Nature Research Editing Service 와 American Journal Experts 에서 저희와 협약을 통해 서비스를 제공하고 있습니다. SpringerOpen에서는 위의 두 가지의 서비스를 첫 논문 투고를 위해 사용하시는 경우, 10%의 할인을 제공하고 있습니다. Nature Research Editing Service이용시 10% 할인을 요청하기 위해서는 여기 를 클릭해 주시고, American Journal Experts 이용시 10% 할인을 요청하기 위해서는 여기 를 클릭해 주십시오.
영문 교정 서비스는 게재를 위한 요구사항은 아니며, 해당 서비스의 이용이 피어 리뷰에 논문이 선택되거나 게재가 수락되는 것을 의미하거나 보장하지 않습니다.
Data and materials
For all journals, SpringerOpen strongly encourages all datasets on which the conclusions of the manuscript rely to be either deposited in publicly available repositories (where available and appropriate) or presented in the main paper or additional supporting files, in machine-readable format (such as spread sheets rather than PDFs) whenever possible. Please see the list of recommended repositories in our editorial policies.
For some journals, deposition of the data on which the conclusions of the manuscript rely is an absolute requirement. Please check the Instructions for Authors for the relevant journal and article type for journal specific policies.
For all manuscripts, information about data availability should be detailed in an ‘Availability of data and materials’ section. For more information on the content of this section, please see the Declarations section of the relevant journal’s Instruction for Authors. For more information on SpringerOpen's policies on data availability, please see our editorial policies .
Formatting the 'Availability of data and materials' section of your manuscript
The following format for the 'Availability of data and materials section of your manuscript should be used:
"The dataset(s) supporting the conclusions of this article is(are) available in the [repository name] repository, [unique persistent identifier and hyperlink to dataset(s) in http:// format]."
The following format is required when data are included as additional files:
"The dataset(s) supporting the conclusions of this article is(are) included within the article (and its additional file(s))."
For databases, this section should state the web/ftp address at which the database is available and any restrictions to its use by non-academics.
For software, this section should include:
- Project name: e.g. My bioinformatics project
- Project home page: e.g. http://sourceforge.net/projects/mged
- Archived version: DOI or unique identifier of archived software or code in repository (e.g. enodo)
- Operating system(s): e.g. Platform independent
- Programming language: e.g. Java
- Other requirements: e.g. Java 1.3.1 or higher, Tomcat 4.0 or higher
- License: e.g. GNU GPL, FreeBSD etc.
- Any restrictions to use by non-academics: e.g. licence needed
Information on available repositories for other types of scientific data, including clinical data, can be found in our editorial policies .
What should be cited?
Only articles, clinical trial registration records and abstracts that have been published or are in press, or are available through public e-print/preprint servers, may be cited.
Unpublished abstracts, unpublished data and personal communications should not be included in the reference list, but may be included in the text and referred to as "unpublished observations" or "personal communications" giving the names of the involved researchers. Obtaining permission to quote personal communications and unpublished data from the cited colleagues is the responsibility of the author. Either footnotes or endnotes are permitted. Journal abbreviations follow Index Medicus/MEDLINE.
Any in press articles cited within the references and necessary for the reviewers' assessment of the manuscript should be made available if requested by the editorial office.
Preparing figures
When preparing figures, please follow the formatting instructions below.
- Figure titles (max 15 words) and legends (max 300 words) should be provided in the main manuscript, not in the graphic file.
- Tables should NOT be submitted as figures but should be included in the main manuscript file.
- Multi-panel figures (those with parts a, b, c, d etc.) should be submitted as a single composite file that contains all parts of the figure.
- Figures should be numbered in the order they are first mentioned in the text, and uploaded in this order.
- Figures should be uploaded in the correct orientation.
- Figure keys should be incorporated into the graphic, not into the legend of the figure.
- Each figure should be closely cropped to minimize the amount of white space surrounding the illustration. Cropping figures improves accuracy when placing the figure in combination with other elements when the accepted manuscript is prepared for publication on our site. For more information on individual figure file formats, see our detailed instructions.
- Individual figure files should not exceed 10 MB. If a suitable format is chosen, this file size is adequate for extremely high quality figures.
- Please note that it is the responsibility of the author(s) to obtain permission from the copyright holder to reproduce figures (or tables) that have previously been published elsewhere. In order for all figures to be open access, authors must have permission from the rights holder if they wish to include images that have been published elsewhere in non open access journals. Permission should be indicated in the figure legend, and the original source included in the reference list.
Figure file types
We accept the following file formats for figures:
- EPS (suitable for diagrams and/or images)
- PDF (suitable for diagrams and/or images)
- Microsoft Word (suitable for diagrams and/or images, figures must be a single page)
- PowerPoint (suitable for diagrams and/or images, figures must be a single page)
- TIFF (suitable for images)
- JPEG (suitable for photographic images, less suitable for graphical images)
- PNG (suitable for images)
- BMP (suitable for images)
- CDX (ChemDraw - suitable for molecular structures)
Figure size and resolution
Figures are resized during publication of the final full text and PDF versions to conform to the SpringerOpen standard dimensions, which are detailed below.
Figures on the web:
- width of 600 pixels (standard), 1200 pixels (high resolution).
Figures in the final PDF version:
- width of 85 mm for half page width figure
- width of 170 mm for full page width figure
- maximum height of 225 mm for figure and legend
- image resolution of approximately 300 dpi (dots per inch) at the final size
Figures should be designed such that all information, including text, is legible at these dimensions. All lines should be wider than 0.25 pt when constrained to standard figure widths. All fonts must be embedded.
Figure file compression
Vector figures should if possible be submitted as PDF files, which are usually more compact than EPS files.
- TIFF files should be saved with LZW compression, which is lossless (decreases file size without decreasing quality) in order to minimize upload time.
- JPEG files should be saved at maximum quality.
- Conversion of images between file types (especially lossy formats such as JPEG) should be kept to a minimum to avoid degradation of quality.
If you have any questions or are experiencing a problem with figures, please contact the customer service team at [email protected] .
Preparing tables
When preparing tables, please follow the formatting instructions below.
- Tables should be numbered and cited in the text in sequence using Arabic numerals (i.e. Table 1, Table 2 etc.).
- Tables less than one A4 or Letter page in length can be placed in the appropriate location within the manuscript.
- Tables larger than one A4 or Letter page in length can be placed at the end of the document text file. Please cite and indicate where the table should appear at the relevant location in the text file so that the table can be added in the correct place during production.
- Larger datasets, or tables too wide for A4 or Letter landscape page can be uploaded as additional files. Please see [below] for more information.
- Tabular data provided as additional files can be uploaded as an Excel spreadsheet (.xls ) or comma separated values (.csv). Please use the standard file extensions.
- Table titles (max 15 words) should be included above the table, and legends (max 300 words) should be included underneath the table.
- Tables should not be embedded as figures or spreadsheet files, but should be formatted using ‘Table object’ function in your word processing program.
- Color and shading may not be used. Parts of the table can be highlighted using superscript, numbering, lettering, symbols or bold text, the meaning of which should be explained in a table legend.
- Commas should not be used to indicate numerical values.
If you have any questions or are experiencing a problem with tables, please contact the customer service team at [email protected] .
Preparing additional files
As the length and quantity of data is not restricted for many article types, authors can provide datasets, tables, movies, or other information as additional files.
All Additional files will be published along with the accepted article. Do not include files such as patient consent forms, certificates of language editing, or revised versions of the main manuscript document with tracked changes. Such files, if requested, should be sent by email to the journal’s editorial email address, quoting the manuscript reference number.
Results that would otherwise be indicated as "data not shown" should be included as additional files. Since many web links and URLs rapidly become broken, SpringerOpen requires that supporting data are included as additional files, or deposited in a recognized repository. Please do not link to data on a personal/departmental website. Do not include any individual participant details. The maximum file size for additional files is 20 MB each, and files will be virus-scanned on submission. Each additional file should be cited in sequence within the main body of text.
Submit manuscript
- Editorial Board
- Sign up for article alerts and news from this journal
- Follow us on Twitter
Affiliated with

Annual Journal Metrics
2023 Speed 14 days submission to first editorial decision for all manuscripts (Median) 96 days submission to accept (Median)
2023 Usage 167,495 downloads 29 Altmetric mentions
- More about our metrics
- ISSN: 2364-9534 (electronic)
- About this blog
- PhD Resources
- Writing a Mini-Review: A Crucial Task in PhD Research
Introduction
One of the research tasks that’s given me a tremendous amount of benefit during my PhD is the writing of a mini-review. I use the term “mini-review” only because this task is something akin to completing a book review (only a little smaller)…and because frankly, I don’t know what else to call what I’m doing.
What is a mini-review
A mini-review (like a book review) is simply the careful summary of a particular scholar’s work. It is usually a 2-3 page articulation of a work’s main thesis, supporting arguments and major points.
After this is complete, a final step in the mini-review is to take the content of 1-2 pages and shrink that down to one paragraph that you place at the top as a summary of the whole review.
Of course, creating a mini-review implies that you’ve actually sat down and spent some time in the book or article. While it may take some time to put it together, the rewards are many (as I will enumerate later on.)
The Why of a Mini-Review
There are at least two major reasons for spending some time on this task:
1) You avoid the temptation to skim a work
Let’s face it, it is often very tempting to superficially skim a work, perhaps combine 2-3 book reviews and think you have an idea of an author’s contribution to your thesis.
It doesn’t take great skill to pull this off. It is quite a different matter to really wrestle with an author’s argument and to reflect on how it fits within your overall thesis.
2) You develop the critical skill of summarizing
There are two skills that are absolutely essentially to the completion of your PhD (these aren’t the only two, but they are pretty important).
One is the ability to summarize an argument the other is the ability to synthesize various works into a cohesive narrative.
One entire chapter of your dissertation (your literature review) is essentially the concise summation (and synthesis) of dozens and dozens of works related to your topic.
But beyond that, every section of your dissertation is an interaction of your ideas with the ideas of other scholars (whose work you must summarize…) Ditto for being able to create rich and meaningful footnotes that capture the essence of a work.
We might even say that summarizing arguments is the work horse of your PhD program around which you create your original contribution.
Benefits of Doing Mini-Reviews
The benefits to doing this kind of leg work early on in your research, and as you work your way through the dissertation are many:
1) You will be completing work for your literature review
Simply take your summary of your mini-review, the final step I mentioned above under “What is a Mini-Review” and you’ve got a pretty good entry to fit somewhere in your literature review.
2) Create expanded bibliographies to provide to your supervisor
Prior to my meetings with my supervisor I would often create a YTD summary of my readings for the time period between our chats. This document often served as a springboard to many of our discussions and it gave my supervisor a quick glance into the scholarship related to my topic.
3) When you sit down to write you will have a wealth of material to draw upon
It is one thing to draw upon someone else’s book review to try to fill in some gaps in your dissertation. It is quite another to have a 1-2 page summary of a work that you have labored to create.
Not only is your knowledge of an author’s work personal and deep, which allows you to have meaningful interactions, but also, your ability to draw connections (both to your thesis and to other related works) is greatly enhanced after completing a mini-review.
4) Your mini-reviews are a great resource for new insights or memory refreshers
Reading your own mini-review is like reading the most salient set of cliff notes on a particular work. If it’s been a while since you’ve picked up a particular work, your mini-review will bring it all to mind.
In addition, reading my mini-reviews has sometimes sparked new ideas and fresh insights for my dissertation if only because some time may have elapsed since the original reading / creation of the book / mini-review.
In the interim, my knowledge and maturity about my topic may have shifted, allowing a fresh re-reading of my mini-review.
5) Writing a mini-review gets you into the groove of writing
If there is one regret I’ve had as I’ve worked on my dissertation it’s been that I’ve done too much research (note-taking) and not enough writing.
While gathering and collating sources is important, the key to a dissertation is output, and the only way of getting output is to slog your way through it.
Mini-reviews get you in the PhD mode. They get you thinking like someone in the academy.
How do you say something concisely, how do you accurately reflect others’ views, what is important and what is not, is this a good argument or not, etc.
A Sample Mini-Review
Click on the link below to see a sample mini-review I created for an article on the Miletus Speech
Lambrecht: Paul’s Farewell Address
While the creation of mini-reviews may take some time, there is often no substitute for an honest wrestling with a particular author’s work.
Endeavoring in such labor, however, pays off rich dividends in your personal knowledge and in the wealth of source material that will be available for different parts of your dissertation.
Happy researching!
3 Responses to Writing a Mini-Review: A Crucial Task in PhD Research
Pingback: Sample Mini-Review – “Paul’s Farewell” by Lambrecht | Phd Tips and Dissertation Advice
Our Children may perform better in school and feel more confident about themselves if they are told that failure is a normal part of learning, rather than being pressured to succeed at all costs, according to new research published by the American Psychological Association.
Pingback: Annotated Bibliography: Why You Should Prepare One | Phd Tips and Dissertation Advice
Comments are closed.
How to Prepare / Succeed in a PhD Program
Download our Guide! Top 10 Dissertation Writing Tips

- Search for:
- Application
- Applying to PhD Program
- Arguments in a Thesis
- Bibliography
- Bibliography Software
- Dissertation Proposal
- Dissertation Topic
- Finding a Supervisor
- Organization
- PhD Resource – Book Review
- Preparation
- Software Tips
- Uncategorized
Top Posts & Pages
- PhD Tips: Note-taking Software for Writing a PhD
- Sample Mini-Review - "Paul's Farewell" by Lambrecht

- Category: AI
Tiny but mighty: The Phi-3 small language models with big potential
- Sally Beatty

Sometimes the best way to solve a complex problem is to take a page from a children’s book. That’s the lesson Microsoft researchers learned by figuring out how to pack more punch into a much smaller package.
Last year, after spending his workday thinking through potential solutions to machine learning riddles, Microsoft’s Ronen Eldan was reading bedtime stories to his daughter when he thought to himself, “how did she learn this word? How does she know how to connect these words?”
That led the Microsoft Research machine learning expert to wonder how much an AI model could learn using only words a 4-year-old could understand – and ultimately to an innovative training approach that’s produced a new class of more capable small language models that promises to make AI more accessible to more people.
Large language models (LLMs) have created exciting new opportunities to be more productive and creative using AI. But their size means they can require significant computing resources to operate.
While those models will still be the gold standard for solving many types of complex tasks, Microsoft has been developing a series of small language models (SLMs) that offer many of the same capabilities found in LLMs but are smaller in size and are trained on smaller amounts of data.
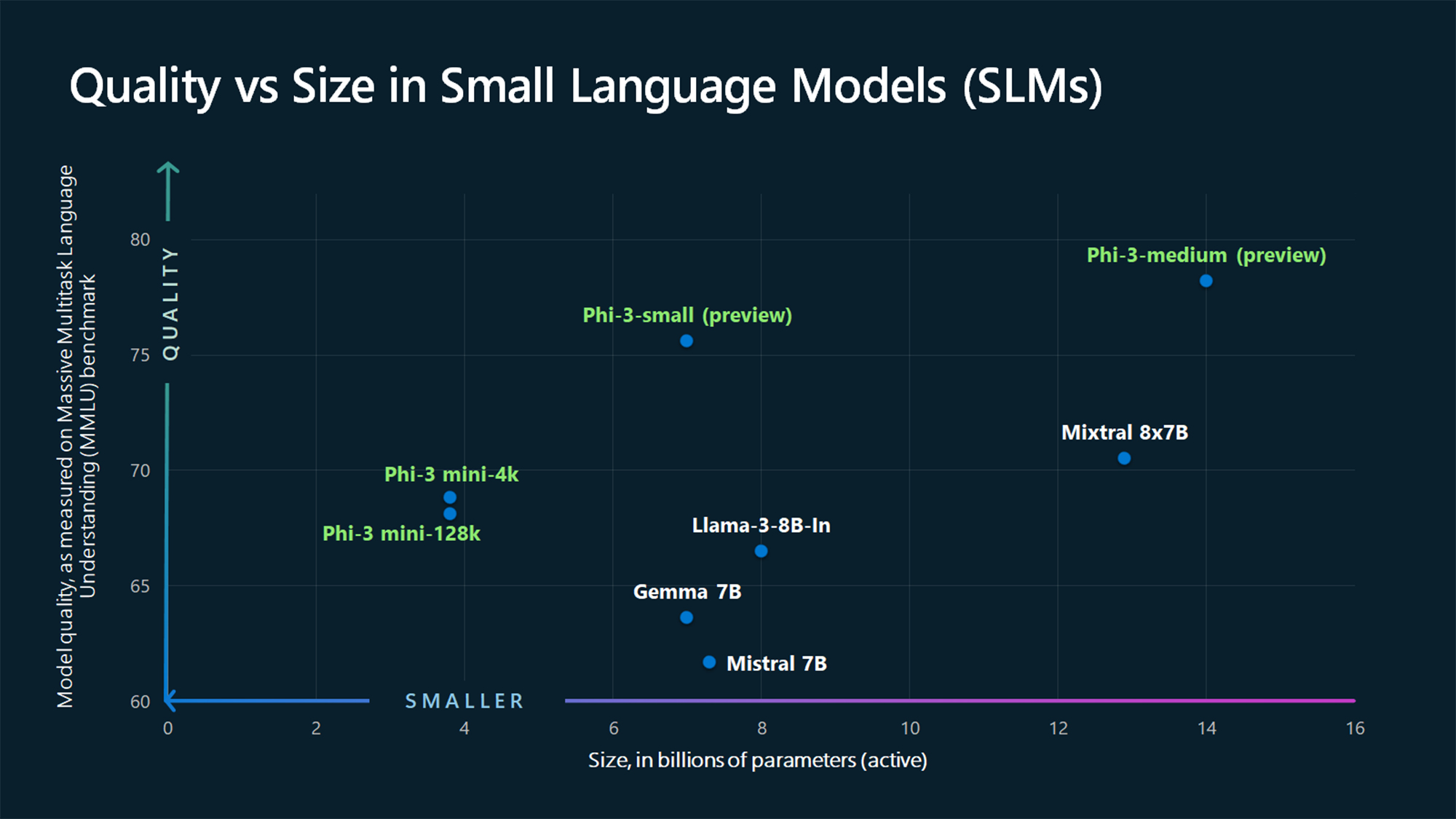
The company announced today the Phi-3 family of open models , the most capable and cost-effective small language models available. Phi-3 models outperform models of the same size and next size up across a variety of benchmarks that evaluate language, coding and math capabilities, thanks to training innovations developed by Microsoft researchers.
Microsoft is now making the first in that family of more powerful small language models publicly available: Phi-3-mini , measuring 3.8 billion parameters, which performs better than models twice its size, the company said.
Starting today, it will be available in the Microsoft Azure AI Model Catalog and on Hugging Face , a platform for machine learning models, as well as Ollama , a lightweight framework for running models on a local machine. It will also be available as an NVIDIA NIM microservice with a standard API interface that can be deployed anywhere.
Microsoft also announced additional models to the Phi-3 family are coming soon to offer more choice across quality and cost. Phi-3-small (7 billion parameters) and Phi-3-medium (14 billion parameters) will be available in the Azure AI Model Catalog and other model gardens shortly.
Small language models are designed to perform well for simpler tasks, are more accessible and easier to use for organizations with limited resources and they can be more easily fine-tuned to meet specific needs.
“What we’re going to start to see is not a shift from large to small, but a shift from a singular category of models to a portfolio of models where customers get the ability to make a decision on what is the best model for their scenario,” said Sonali Yadav, principal product manager for Generative AI at Microsoft.
“Some customers may only need small models, some will need big models and many are going to want to combine both in a variety of ways,” said Luis Vargas, vice president of AI at Microsoft.
Choosing the right language model depends on an organization’s specific needs, the complexity of the task and available resources. Small language models are well suited for organizations looking to build applications that can run locally on a device (as opposed to the cloud) and where a task doesn’t require extensive reasoning or a quick response is needed.
Large language models are more suited for applications that need orchestration of complex tasks involving advanced reasoning, data analysis and understanding of context.
Small language models also offer potential solutions for regulated industries and sectors that encounter situations where they need high quality results but want to keep data on their own premises, said Yadav.
Vargas and Yadav are particularly excited about the opportunities to place more capable SLMs on smartphones and other mobile devices that operate “at the edge,” not connected to the cloud. (Think of car computers, PCs without Wi-Fi, traffic systems, smart sensors on a factory floor, remote cameras or devices that monitor environmental compliance.) By keeping data within the device, users can “minimize latency and maximize privacy,” said Vargas.
Latency refers to the delay that can occur when LLMs communicate with the cloud to retrieve information used to generate answers to users prompts. In some instances, high-quality answers are worth waiting for while in other scenarios speed is more important to user satisfaction.
Because SLMs can work offline, more people will be able to put AI to work in ways that haven’t previously been possible, Vargas said.
For instance, SLMs could also be put to use in rural areas that lack cell service. Consider a farmer inspecting crops who finds signs of disease on a leaf or branch. Using a SLM with visual capability, the farmer could take a picture of the crop at issue and get immediate recommendations on how to treat pests or disease.
“If you are in a part of the world that doesn’t have a good network,” said Vargas, “you are still going to be able to have AI experiences on your device.”
The role of high-quality data
Just as the name implies, compared to LLMs, SLMs are tiny, at least by AI standards. Phi-3-mini has “only” 3.8 billion parameters – a unit of measure that refers to the algorithmic knobs on a model that help determine its output. By contrast, the biggest large language models are many orders of magnitude larger.
The huge advances in generative AI ushered in by large language models were largely thought to be enabled by their sheer size. But the Microsoft team was able to develop small language models that can deliver outsized results in a tiny package. This breakthrough was enabled by a highly selective approach to training data – which is where children’s books come into play.
To date, the standard way to train large language models has been to use massive amounts of data from the internet. This was thought to be the only way to meet this type of model’s huge appetite for content, which it needs to “learn” to understand the nuances of language and generate intelligent answers to user prompts. But Microsoft researchers had a different idea.
“Instead of training on just raw web data, why don’t you look for data which is of extremely high quality?” asked Sebastien Bubeck, Microsoft vice president of generative AI research who has led the company’s efforts to develop more capable small language models. But where to focus?
Inspired by Eldan’s nightly reading ritual with his daughter, Microsoft researchers decided to create a discrete dataset starting with 3,000 words – including a roughly equal number of nouns, verbs and adjectives. Then they asked a large language model to create a children’s story using one noun, one verb and one adjective from the list – a prompt they repeated millions of times over several days, generating millions of tiny children’s stories.
They dubbed the resulting dataset “TinyStories” and used it to train very small language models of around 10 million parameters. To their surprise, when prompted to create its own stories, the small language model trained on TinyStories generated fluent narratives with perfect grammar.
Next, they took their experiment up a grade, so to speak. This time a bigger group of researchers used carefully selected publicly-available data that was filtered based on educational value and content quality to train Phi-1. After collecting publicly available information into an initial dataset, they used a prompting and seeding formula inspired by the one used for TinyStories, but took it one step further and made it more sophisticated, so that it would capture a wider scope of data. To ensure high quality, they repeatedly filtered the resulting content before feeding it back into a LLM for further synthesizing. In this way, over several weeks, they built up a corpus of data large enough to train a more capable SLM.
“A lot of care goes into producing these synthetic data,” Bubeck said, referring to data generated by AI, “looking over it, making sure it makes sense, filtering it out. We don’t take everything that we produce.” They dubbed this dataset “CodeTextbook.”
The researchers further enhanced the dataset by approaching data selection like a teacher breaking down difficult concepts for a student. “Because it’s reading from textbook-like material, from quality documents that explain things very, very well,” said Bubeck, “you make the task of the language model to read and understand this material much easier.”
Distinguishing between high- and low-quality information isn’t difficult for a human, but sorting through more than a terabyte of data that Microsoft researchers determined they would need to train their SLM would be impossible without help from a LLM.
“The power of the current generation of large language models is really an enabler that we didn’t have before in terms of synthetic data generation,” said Ece Kamar, a Microsoft vice president who leads the Microsoft Research AI Frontiers Lab, where the new training approach was developed.
Starting with carefully selected data helps reduce the likelihood of models returning unwanted or inappropriate responses, but it’s not sufficient to guard against all potential safety challenges. As with all generative AI model releases, Microsoft’s product and responsible AI teams used a multi-layered approach to manage and mitigate risks in developing Phi-3 models.
For instance, after initial training they provided additional examples and feedback on how the models should ideally respond, which builds in an additional safety layer and helps the model generate high-quality results. Each model also undergoes assessment, testing and manual red-teaming, in which experts identify and address potential vulnerabilities.
Finally, developers using the Phi-3 model family can also take advantage of a suite of tools available in Azure AI to help them build safer and more trustworthy applications.
Choosing the right-size language model for the right task
But even small language models trained on high quality data have limitations. They are not designed for in-depth knowledge retrieval, where large language models excel due to their greater capacity and training using much larger data sets.
LLMs are better than SLMs at complex reasoning over large amounts of information due to their size and processing power. That’s a function that could be relevant for drug discovery, for example, by helping to pore through vast stores of scientific papers, analyze complex patterns and understand interactions between genes, proteins or chemicals.
“Anything that involves things like planning where you have a task, and the task is complicated enough that you need to figure out how to partition that task into a set of sub tasks, and sometimes sub-sub tasks, and then execute through all of those to come with a final answer … are really going to be in the domain of large models for a while,” said Vargas.
Based on ongoing conversations with customers, Vargas and Yadav expect to see some companies “offloading” some tasks to small models if the task is not too complex.

For instance, a business could use Phi-3 to summarize the main points of a long document or extract relevant insights and industry trends from market research reports. Another organization might use Phi-3 to generate copy, helping create content for marketing or sales teams such as product descriptions or social media posts. Or, a company might use Phi-3 to power a support chatbot to answer customers’ basic questions about their plan, or service upgrades.
Internally, Microsoft is already using suites of models, where large language models play the role of router, to direct certain queries that require less computing power to small language models, while tackling other more complex requests itself.
“The claim here is not that SLMs are going to substitute or replace large language models,” said Kamar. Instead, SLMs “are uniquely positioned for computation on the edge, computation on the device, computations where you don’t need to go to the cloud to get things done. That’s why it is important for us to understand the strengths and weaknesses of this model portfolio.”
And size carries important advantages. There’s still a gap between small language models and the level of intelligence that you can get from the big models on the cloud, said Bubeck. “And maybe there will always be a gap because you know – the big models are going to keep making progress.”
Related links:
- Read more: Introducing Phi-3, redefining what’s possible with SLMs
- Learn more: Azure AI
- Read more: Phi-3 Technical Report: A Highly Capable Language Model Locally on Your Phone
Top image: Sebastien Bubeck, Microsoft vice president of Generative AI research who has led the company’s efforts to develop more capable small language models. (Photo by Dan DeLong for Microsoft)
Advertisement
Supported by
‘The New Edition Story’: A Loving Tribute to a Talented but Tormented Group
- Share full article

By Jon Caramanica
- Jan. 23, 2017
Of all forms of homage, attention to detail is perhaps the most loving. It requires not just knowledge and dedication, but also thoughtfulness. It prizes accuracy both out of respect to the historical record, but also to those who wrote it. It celebrates the vision of the original creators more than the disruptive instincts of reinterpreters.
By that measurement, “The New Edition Story,” a vibrant, fiercely committed three-night mini-series that begins Tuesday on BET, is overflowing with love — a jubilant celebration of a group that was preternaturally talented and rivetingly tortured.
Few groups in modern black pop are as deserving of this treatment as New Edition, which transported the elegance of the male vocal groups of the 1970s into the first wave of R&B’s dance with hip-hop in the late 1980s . The act released four essential albums, a couple more good ones, and spun off careers for all its members — solo work by Bobby Brown, Johnny Gill and Ralph Tresvant, and a group, Bell Biv DeVoe, made up of the others.
None of this happened without conflict. New Edition, which hailed largely from the Orchard Park projects in the Roxbury section of Boston, was a black act that was chided by its label for not being mainstream enough. The members squabbled with executives, management and among themselves. This was in many ways the archetype of the modern boy band — so much so that when New Edition left Maurice Starr, who produced the group’s early songs, he created New Kids on the Block, essentially a white copy.
For all of its musical influence, the group is probably best known for its internal dramas, its contractual nightmares and for the troubled adulthood of Mr. Brown. “The New Edition Story” isn’t hagiography. Friction was integral to the group’s mystique, but the show spends a good deal of time on its earliest days as preteens trying to forge a style. (For this part of the series, the members are played by younger actors. Tyler Williams as Mr. Brown and Jahi Winston as Mr. Tresvant are particularly dynamic).
New Edition’s ascent was rapid: “Candy Girl,” the group’s 1983 debut single, went to No. 1 on the Billboard hot black singles chart, and the group had successful hits until the end of the decade. But in this mini-series, that stretch of time is depicted as one of turmoil and dissatisfaction. Mr. Tresvant (Algee Smith, tender and limber) carries the disproportionate burdens of the lead singer, earning acclaim and resentment. And of course, there’s Mr. Brown (a combustible Woody McClain), who’s perpetually at war: with the other group members, with management, with law enforcement and, of course, with himself .
“The New Edition Story” depicts a group of young men consistently slipping through the fingers of authority figures. That means their mothers (Sandi McCree as Carole Brown and Yvette Nicole Brown as Shirley Bivins are especially intense); their manager and choreographer Brooke Payne, played by Wood Harris, who conveys a world-weary swagger with the tiniest cock of the head or extinguishing of a cigarette; and their manager (in this case Gary Evans, a fictional stand-in for the group’s manager in its late 1980s pop heyday), played by a manic Michael Rapaport.
“The New Edition Story” is, by far, the best of the recent spate of black pop biopics, miles beyond the shoestring Lifetime entries about Toni Braxton and Aaliyah, films that have contributed to an air of lowered expectations for projects of this nature. Instead, “The New Edition Story” — written by Abdul Williams and directed by Chris Robinson — is more in keeping with the excellent “Unsung” docu-series about black musicians on TV One. It tells the group’s story, warts and all. (But not all the warts: Many of the details of Mr. Brown’s tumult have been left aside, and certain incidents, such as the behind-the-scenes late-80s quarrels with the R&B group Guy that led to the shooting death of that group’s head of security, didn’t make the cut.)
It undoubtedly helps the mini-series that the whole group — the original members Mr. Brown, Michael Bivins, Mr. Tresvant, Ricky Bell and Ronnie DeVoe, as well as Mr. Gill, who joined after Mr. Brown’s departure — signed on as co-producers. While that might mean some light sugarcoating, it also results in a historical narrative told with impressive detai l, particularly in regards to wardrobe and production design (kudos to the costume designer Rita McGhee). Album covers, dance routines and video shoots are remade down to the tiniest details (the remade videos for “If It Isn’t Love” and Mr. Brown’s “Every Little Step” are impressive). The clothing is a virtuosic symphony of sequined suits, flowing synthetic fabrics, graffitied overalls.
And then there’s the music: “The New Edition Story” relies on effective rerecorded versions of the group’s many hits, sung by the actors (and mixed in places with original master recordings.) The music itself is handled by the duos of Babyface and Antonio Dixon, and Jimmy Jam and Terry Lewis, who’ve rebuilt the New Edition sound (much of which they originally created) with rigor and affection.
Approval and faithfulness have limits, though. The group’s founding is streamlined, and some later-era slang slips into early-era scenes. Comparatively little time is spent on the group members’ solo efforts, especially given that so much of the final installment focuses on the members’ reconciliation in the mid-2000s, a reassurance that everyone ended up on the same page, putting their individual and collective troubles behind them. (The N.W.A biopic “Straight Outta Compton” took a similar, though more maudlin approach.)
Here, some liberties have been taken with the timeline, placing an impromptu reunion at Mr. DeVoe’s wedding, in 2006, before the group’s participation at a BET 25th anniversary show, in 2005, so that the mini-series can effectively end with a greatest-hits performance and a BET infomercial. This is slick positioning and salesmanship, but also disingenuous. New Edition has been as fascinating for its valleys as its peaks. For this group, the most fitting tribute is always the truth.
The New Edition Story A mini-series starting Tuesday on BET.
Explore More in TV and Movies
Not sure what to watch next we can help..
Sydney Sweeney and Glen Powell speak about how “Anyone but You” beat the rom-com odds. Here are their takeaways after the film , debuting on Netflix, went from box office miss to runaway hit.
The vampire ballerina in the new movie “Abigail” has a long pop culture lineage . She and her sisters are obsessed, tormented and likely to cause harm.
In a joint interview, the actors Lily Gladstone and Riley Keough discuss “Under the Bridge,” their new true-crime series based on a teenager’s brutal killing in British Columbia.
The movie “Civil War” has tapped into a dark set of national angst . In polls and in interviews, a segment of voters say they fear the country’s divides may lead to actual, not just rhetorical, battles.
If you are overwhelmed by the endless options, don’t despair — we put together the best offerings on Netflix , Max , Disney+ , Amazon Prime and Hulu to make choosing your next binge a little easier.
Sign up for our Watching newsletter to get recommendations on the best films and TV shows to stream and watch, delivered to your inbox.
Drug Addiction Issues in The Corner Miniseries Research Paper
Introduction, body of the paper.
Nowadays, it became a commonplace practice among many people to refer to the problem of drug-addiction, as one of the most pressing social issues in today’s America. This state of affairs appears thoroughly justified. After all, it does not represent much of a secret that, as of today, just about every large American city features the so-called ‘bad areas’ (believed to be swarmed with drug-addicts), where socially upstanding residents try not to venture, unless absolutely necessary – just as it is being shown in the HBO miniseries The Corner (2000).
What adds to the sheer acuteness of the issue in question is that the currently deployed anti-drug policies, mostly concerned with the functioning of America’s justice system, can hardly be deemed very effective. One the reasons for this is that most of these policies do not take into account the provisions of the Ecological Systems Theory EST (developed by Urie Bronfenbrenner), which stresses out the importance of environmental factors of influence, within the context of how people go about constructing their sense of self-identity and choosing in favor one or another behavioral pattern.
As Duerden and Witt defined it: “EST proposes that individuals exist within a variety of settings, starting at the individual level and extending outward (e.g., family, work, society, etc.)… (deems) development as a process that involves interactions both within and across contexts” (2010, p. 109). In my paper, I will explore the validity of this suggestion at length, in regards to the themes and motifs, contained in The Corner , while promoting the idea that the realities of a post-industrial living in America call for the reassessment of the very methodological approach to conducting the ‘war on drugs’.
Probably the main reason why The Corner does deserve to be considered utterly insightful, in the sense of how it treats the theme of drug-addiction, is that it exposes the fallaciousness of the assumption that one’s addiction is best discussed within the context of what happened to be the addicted person’s ethno-cultural/racial affiliation. After all, even though most of African-American characters, featured the miniseries, are indeed shown struggling with their drug-addictions, there appears to have been nothing biologically deterministic about how these individuals ended up being ‘hooked on drugs’.
The character of Gary McCullough comes in as a good example, in this respect. Despite the fact that Gary’s addiction seems to have been embedded into his personality from the very beginning, in the Episode 3 we learn that this character was once a bright college student, who wanted to become a stock-market broker. The fact that, throughout the series’ entirety, Gary never ceases to exhibit his talent in carpentry, also suggests that, if anything, he is the least deserves to be referred to as a ‘natural born’ drug-addict.
Essentially same can be said about every other character in The Corner – despite being used to drugs and to the drug-related street violence, they nevertheless continue to exhibit their full awareness that leading the lifestyle of a drug-addict is utterly inappropriate. Moreover, many of them make a deliberate point in pledging to raise their children in the drug-free environment. To exemplify the validity of this suggestion, we can refer to the characters of DeAndre (Gary’s son) and Tyreeka (DeAndre’s girlfriend).
After having found out that Tyreeka was pregnant with his baby, DeAndre declares that he will do just about all it takes, in order to make sure that his child would be spared from having to learn the ways of the street (Episode 5). In her turn, Tyreeka proves herself thoroughly aware that the notion of parenthood is synonymous with the notion of responsibility: “I do feel like I’m ready to be a mother, because I will give my baby lots of love and my baby is going to love me back” (00.56.03).
When assessed through the conceptual lenses of EST, the above-mentioned observations can be interpreted as the indication that, contrary to what it is being commonly assumed, the identity of being African-Americans did not have any direct link to these characters’ affiliation with drug/gangsta culture. The reason for this is that, as it can be seen in the series, the micro-systemic (family-related) aspects of one’s upbringing in the drug-infested ethnic ‘ghetto’, do not necessarily presuppose that the concerned individual would be naturally inclined to experiment with drugs. Quite on the contrary – due to having been exposed to the effects of a drug-abuse, ever since its early childhood-years, he or she should be innately resentful of the idea.
What it means is that the actual answer, as to why African-American culture continues to be considered ‘drug-infested’, should be sought at the higher levels of what EST conceptualizes as the process of a socially integrated individual striving to attain self-actualization. According to the theory, the process’s sub-sequential phase is concerned with the influence, exerted upon a person, by the peculiarities of how he or she goes about trying to socialize with others, within the same socio-environmental niche, which EST refers to in terms of ‘mesosystem’.
As Bronfenbrenner described it: “Mesosystem is… a place where people can readily engage in face-to-face interaction” (197, p. 22). As it can be seen in The Corner , it is specifically while being the part of the ‘Crenshaw mafia’ (consisting of his closest friends), that DeAndre became perceptually and cognitively tolerant towards the idea that there is nothing wrong about peddling drugs out on the street. The reason for this is that, while being a young male, DeAndre naturally strived to attain a dominant status among its peers, which in turn could be accomplished by the mean of positioning himself as a ‘tough’ but thoroughly rational individual, who addresses life-challenges in the most energetically sound manner.
For example, upon having been asked (Episode 2) whether he experiences any remorse for the fact that, while peddling drugs, he in fact helps his ‘clients’ to destroy their lives, DeAndre replies: “People who use, they go use. They go buy it from somebody somewhere. Might as well be me” (00.02.27). This DeAndre’s statement provides us with the insight into the workings of his psyche, as such that have been shaped by the realities of living in one of West Baltimore’s ‘hoods’ – the place where, due to being concerned with trying to survive, people could not care less about acting morally. What this means is that DeAndre’s stance in life can be discussed as having been partially reflective of what were the specifics of the peer-pressure in the neighborhood, which he never ceased experiencing.
Essentially the same can be said about many of the series’ other prominent characters, such Gary. The reason for this is that many of this character’s relapses back to using drugs, can be explained by the fact that, throughout the course of all six episodes, he never declines to offer a helping hand to those of his drug-addicted friends that happened to be in need. Unfortunately, most of the time this ‘help’ was concerned with Gary giving his friends money to buy heroin. It is understood, of course, that this could not result in anything else but in keeping Gary in a ‘drug-tolerant’ mood – hence, weakening his desire to put away with the addiction.
Nevertheless, even though that, when assessed from the methodological perspective of EST, many of the series’ themes and motifs do appear to be rather ‘mesosystemic’, there can be only a few doubts as to the overall ‘marcrosystemic’ sounding of The Corner . The reason for this is that, as the series imply, the fact that many African-Americans are indeed addicted to drugs, cannot be discussed outside of the discourse of ‘euro-centrism’, which continues to define the qualitative subtleties of how this country actually operates.
This implicit message, conveyed by the series, fully correlates with the theory’s outlook on ‘macrosystem’ as: “the set of social patterns that govern the formation and dissolution of social interactions between individuals, and thus the relationship among ecological systems” (Neal & Neal, 2013, p. 729). In other words, the actual reason why many African-Americans cannot help becoming drug-addicts, is that they continue being explicitly and implicitly discriminated against, in the social sense of this word. In its turn, this situation appears to have been predetermined by the essence of the U.S. economy’s operational principles, on one hand, and by the fact that many American Whites continue to hold prejudices against their African-American co-citizens, on the other.
In The Corner , there are a number of scenes that illustrate the validity of this suggestion. For example, at the beginning of the Episode 2, DeAndre comes up with the statement: “It’s a hard work selling drugs in Baltimore ghetto. It’s a hard work being a Black man in America’ (00.01.58). As this statement implies, it was not that DeAndre chose to become a drug-dealer, because the dubious ‘career’ in question really did appeal to him, but that he was simply trying to survive within the hostile social environment. After all, throughout the series, he never ceases to act as a ‘hunter-gatherer’ for his mother Fran and later for Tyreeka.
This, of course, can be interpreted as yet another proof that EST is indeed legitimate, because many of its conceptual provisions do appear thoroughly consistent with the realities of one’s ‘ghetto-living’, as seen in The Corner . For example, the fact that, despite understanding the dangers of drugs, DeAndre continues to sell them, does support the theory’s convention that the manner in which people tend to act, is being reflective of what happened to be the affiliated social circumstances. In DeAndre’s case, the main of these circumstances was his (and his mother’s) condition of poverty.
However, the clearest indication that the issue of drug-addiction in African-American communities is macro-dimensional, appears to be the scene (Episode 4), in which Gary goes to watch Schindler’s List in the movie-theatre and consequently elaborates on what he considers the actual meaning of what he saw. While talking to his friends (drug-addicts), Gary says: “Germans said to Jews you ain’t humans… In the end, the Germans decided to kill all the Jews, because they couldn’t seem them being any better than bugs or rats…
I and sit there watching this movie, while realizing that this is happening again” (00.47.20). Apparently, in the aftermath of having watched this film, Gary realized that it was not that much of his own fault for having failed repeatedly, while trying to become ‘clean’, than that of the society. The reason for this is that, throughout the series, most of the featured characters suffer from being dehumanized by the authorities. There is another memorable scene in the same Episode, where a White police officer refers to Scalio (one of Gary’s drug-addicted friends) in such a way, as if the latter were not a human being, but some soulless commodity (00.11.36).
One may wonder about how it can be explained that, while proclaiming its intention to defend ‘human rights’ around the world, America could not come up with any better idea, as to how its own growing population of drug-addicts should be dealt with, then treating them as a ‘social burden’? The logic behind this can be outlined as follows:
As The Corner series imply, the truly effective solution to the problem of drug-addiction/trafficking would have to be concerned with overhauling the country’s systems of justice and education – a clearly macroeconomic undertaking. However, the implementation of this undertaking would cost billions of dollars. The anticipated positive effects would only become apparent in the long-term perspective. What is even more – there is no economically justified motive for the Government to be actually trying to win in the ‘war on drugs’.
The reason for this is that, due to the Globalization-induced ‘outsourcing’, concerned with the process of more and more American companies deciding to move their production lines to the Third and Second World countries, America grows progressively incapable of generating any de facto wealth, and not merely ‘wealth-indicating’ liabilities (in the form of stock-market derivatives, bonds, treasuries, securities, etc.). What it means is that, as time goes on, the employed status of many Americans will be increasingly referred to as a mere formality – especially if they happened to specialize in such clearly post-industrial professional pursuits, as management or public relations.
In other words, there is no any objective need in trying to rehabilitate as many drug-addicts, as possible, so that they would be able to become the society’s productive members. After all, the American society itself had ceased being productive, while growing increasingly consumerist. This is exactly the reason why, while trying to become employed, DeAndre could not secure any other, but the so-called ‘dead end’ jobs, concerned with the economy’s servicing sector, such as mopping floors and flipping burgers at Wendy’s.
In other words, had American drug-addicts been helped to put an end to their addiction, they would begin representing even more acute of a ‘social burden’, because once ‘clean’, these people would begin demanding to be allowed to pursue the so-called ‘American dream’ (making good money), just like the rest. In its turn, this would add to the intensity of social tensions in the U.S. Therefore, it makes much more sense for the Government to simply provide drug-addicted citizens with monthly welfare-checks (large enough for the recipients to afford buying drugs), while expecting that these individuals will simply die off quietly in their ‘ghettos’.
Thus, it will only be logical to suggest that, as of today, the U.S. Government does not seem to have any plan, whatsoever, as to how address the problem of drug-addiction of America on any of the mentioned ecological levels. The best proof to the validity of this suggestion can serve the scene (Episode 1), from which we learn about the actual ‘progress’, achieved on the way of America conducting the ‘war on drugs’: “Thirty years ago, Maryland had only five penal institutions. Today, there are twenty eight” (00.01.03). In another memorable scene (Episode 3), upon having been asked the question “are we going to win this war on drugs?”, the Police Officer Robert Brown replies “no comment” (00.55.41).
Therefore, the problem in question is now being primarily tackled at the micro-level. Even though that the deployment this particular strategy by non-governmental organizations/volunteers is often being hampered by the lack of funds, it nevertheless cannot be referred to as anything but thoroughly justified. The fact that this indeed happened to be the case, can be exemplified, in regards to the character of Ella Thompson, who succeeded rather splendidly in endowing many children in West Baltimore with the aversion towards the very idea of trying drugs – doing this was her own initiative.
In my opinion, in order for the problem of drug-addiction in the U.S. to begin becoming progressively less acute, this country’s policy-makers should first familiarize themselves with the macro-systemic magnitude of the concerned subject matter. Then, it should become clear to them that the first thing that would have to be done, in this respect, would be legitimizing the idea that one’s addiction to drugs is more of an illness than of a criminal offense.
This, however, would prove being easier said than done. After all, there can be no doubt that the representatives of the so-called ‘moral majority’ in this country would strongly oppose the suggested initiative. As Yates and Fording pointed out: “State punitiveness does not appear to be driven by governmental responsiveness to mass ideology… the use of imprisonment is tied to the ideological tenor of the elite political environment and politicians’ electoral incentives” (2005, p. 1118).
Yet, it is only after the Government adopts an intellectually flexible approach towards addressing the issue in question, that it may begin becoming progressively more manageable. For those familiar with the main discursive conventions of EST, this suggestion will appear thoroughly reasonable, because it emphasizes the sheer inappropriateness of applying the solely euro-centric standards for assessing the measure of one’s behavioral adequateness, especially if the concerned individual happened to be ethnically visible.
I believe that the deployed line of argumentation, in regards to the discussed subject matter, is fully consistent with the paper’s initial thesis. Apparently, there are indeed a number of reasons to recommend The Corner for watching by those who are interested in learning about what account for the qualitative aspects of how the problem of drug-addiction undermines the integrity of American society, in general, and the people’s hope that it can be successfully dealt with, in particular. The watching of these series will also come in handy for those, who strive to gain a better understanding of how EST can be deployed, when it comes to defining the discursive significance of a particular socially interactive phenomenon.
Bronfenbrenner, U. (1979). The ecology of human development: Experiments by nature and design. Cambridge, MA: Harvard University Press.
Duerden, M. D., & Witt, P. A. (2010). An Ecological systems theory perspective on youth programming. Journal of Park & Recreation Administration , 28 (2), 108-120.
McVries, P. (2014). The Corner Episode 1 VOSTFR_ Gary’s blues [Video file]. Web.
McVries, P. (2015). The Corner Episode 2 VOSTFR_ DeAndre’s blues [Video file]. Web.
McVries, P. (2014). The Corner Episode 3 VOSTFR_ Fran’s blues [Video file]. Web.
McVries, P. (2014). The Corner episode 5 VOSTFR_Corner boy blues [Video file]. Web.
Neal, J. W., & Neal, Z. P. (2013). Nested or networked? Future directions for Ecological Systems Theory. Social Development , 22 (4), 722-737.
Yates, J. & Fording, R. (2005). Politics and state punitiveness in black and white. The Journal of Politics , 67 (4), 1099-1121.
- Chicago (A-D)
- Chicago (N-B)
IvyPanda. (2020, June 30). Drug Addiction Issues in The Corner Miniseries. https://ivypanda.com/essays/drug-addiction-issues-in-the-corner-miniseries/
"Drug Addiction Issues in The Corner Miniseries." IvyPanda , 30 June 2020, ivypanda.com/essays/drug-addiction-issues-in-the-corner-miniseries/.
IvyPanda . (2020) 'Drug Addiction Issues in The Corner Miniseries'. 30 June.
IvyPanda . 2020. "Drug Addiction Issues in The Corner Miniseries." June 30, 2020. https://ivypanda.com/essays/drug-addiction-issues-in-the-corner-miniseries/.
1. IvyPanda . "Drug Addiction Issues in The Corner Miniseries." June 30, 2020. https://ivypanda.com/essays/drug-addiction-issues-in-the-corner-miniseries/.
Bibliography
IvyPanda . "Drug Addiction Issues in The Corner Miniseries." June 30, 2020. https://ivypanda.com/essays/drug-addiction-issues-in-the-corner-miniseries/.
- Gender Propagation in Titanic Miniseries
- Non-Verbal Communication in "When They See Us" TV Show
- Society, Culture, Economy in "Capitalism" Mini-Series
- Drug Legalization and Intellectual Horizons
- Mass Culture's Influence on Attitude to Drugs
- Different Types of Modern Awards Review
- The Issue of American Horror Story
- “Winter Sonata” and “Dae Jang Geum”: An Analysis of Successful Korean Drama
- Bitterness and Cruelty of War: "Dulce Et Decorum Est" and "Facing It"
- Anti-War Owen’s “Dulce et Decorum Est” and Dylan’s “Masters of War”
- Disease Concept of Alcoholism
- Driving Under the Influence of Alcohol
- Alcohol Addiction and Its Societal Influence
- Substance Abuse Disorders and Their Factors
- Employees of Substance Abuse Treatment Facility Needs
Help | Advanced Search
Computer Science > Computation and Language
Title: phi-3 technical report: a highly capable language model locally on your phone.
Abstract: We introduce phi-3-mini, a 3.8 billion parameter language model trained on 3.3 trillion tokens, whose overall performance, as measured by both academic benchmarks and internal testing, rivals that of models such as Mixtral 8x7B and GPT-3.5 (e.g., phi-3-mini achieves 69% on MMLU and 8.38 on MT-bench), despite being small enough to be deployed on a phone. The innovation lies entirely in our dataset for training, a scaled-up version of the one used for phi-2, composed of heavily filtered web data and synthetic data. The model is also further aligned for robustness, safety, and chat format. We also provide some initial parameter-scaling results with a 7B and 14B models trained for 4.8T tokens, called phi-3-small and phi-3-medium, both significantly more capable than phi-3-mini (e.g., respectively 75% and 78% on MMLU, and 8.7 and 8.9 on MT-bench).
Submission history
Access paper:.
- HTML (experimental)
- Other Formats
References & Citations
- Google Scholar
- Semantic Scholar
BibTeX formatted citation
Bibliographic and Citation Tools
Code, data and media associated with this article, recommenders and search tools.
- Institution
arXivLabs: experimental projects with community collaborators
arXivLabs is a framework that allows collaborators to develop and share new arXiv features directly on our website.
Both individuals and organizations that work with arXivLabs have embraced and accepted our values of openness, community, excellence, and user data privacy. arXiv is committed to these values and only works with partners that adhere to them.
Have an idea for a project that will add value for arXiv's community? Learn more about arXivLabs .
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Isr J Health Policy Res

Mini research projects as a mechanism to improve the quality of dementia care
Hava golander.
Department of Nursing and Herczeg Institute for the Study of Aging and Old Age, Tel Aviv University, Tel Aviv, Israel
Associated Data
Not applicable
Several models have been proposed to connect academia and practice in order to improve long-term care. In this paper we propose and describe the “Mini-Research Group” as an alternative model of such collaboration. The formation of mini-research groups was the unplanned by-product of a longitudinal action research project headed by the late Prof. Rebecca Bergman, a prominent nursing leader from the Department of Nursing at Tel-Aviv University. It involved a two-stage project aimed at developing, and later implementing, a specific tool to evaluate the quality of care provided in geropsychiatric units and to design a nursing intervention which entailed an improved model for care in specialized geropsychiatric units for persons with dementia. Initially, this article describes the projects that led to the development of mini-research groups, and then continues to describe several mini-research projects, focusing on the research questions which emerged from practice as well as the variety of methodologies used. Finally, we discuss the ways in which mini- research groups contributed to the quality of care for persons with dementia, benefited their families, professional staff, faculty participants, and advanced policy development. We argue that in light of the present array of ethical and legal restrictions which inhibit the recruitment of participants, using mini-research groups combined of practitioners and researchers, can provide a pragmatic solution, not only to overcome these barriers, but to improve the quality of care, stimulate clinical dementia research, and promote new insights into the lives of persons with dementia.
Several models have been proposed to connect research with practice in order to improve long-term care, among them research institutes affiliated with nursing homes, clinician-initiated research programs, or the more comprehensive tri-focal model of care which combines patient centered care, positive work environment, and evidence-based practice under one big umbrella which fosters a collaborative relationship between nursing homes and academic institutions. [ 1 – 3 ] Despite their prior successes, these models seem to have disappeared from the field of dementia research. This paper sets forth an Israeli model for addressing this and other challenges: the mini-research group.
The initiative to improve and evaluate the effectiveness of care in geropsychiatric units, which was started by Prof. Rebecca Bergman in 1985 and completed in 1992, produced important lessons for understanding persons in advanced stages of dementia and for assessing care provided and research conducted in geropsychiatric units. This longitudinal action research involved about 70 nurses from 20 geriatric centers in Israel, national geriatric inspectors from the Ministry of Health and faculty members from Tel Aviv University. The establishment of the mini-research groups was one of a number of unplanned positive outcomes which emerged from this project [ 4 ].
A previous report of an interdisciplinary committee on “quality of care in services for the elderly” [ 5 ] provided a comprehensive framework for Prof. Bergman’s project. The basic undifferentiated model consisted of six major domains: physical environment, psychological environment, basic personal care, health care, family involvement and human resources. Thus, the first stage of the project involved further developing a specific model which would be relevant and unique to the geropsychiatric units’ characteristics. This involved reviewing the literature, conducting on-site observations, and interviewing residents, families and staff caregivers. The tool that was developed was tested in several settings [ 6 ]. It related to residents as individuals, as groups, and to the unit as a whole. The model provided for 72 cells which evaluated nine focus items on eight administrative, affective, and instrumental measures, as shown in Fig. Fig.1 1 .

A model of the two-dimensional model containing 72 cells
The second phase of the project included the implementation of the tool. The leading project team organized bimonthly full day meetings attended by more than 70 nurses, including unit nurses and directors from 20 geriatric centers, national geriatric nursing inspectors, and nursing faculty from Tel-Aviv University. Each gathering, hosted by a different geriatric center, followed a similar format: presentation of a background paper, discussion of one of the measures of care, guided tours of geropsychiatric units, and exchanges of information regarding problems and experiences related to the topic in discussion. In addition, the project core team, consisting of three geriatric nurse specialists, provided in depth guidance to six non-profit geropsychiatric units during weekly site visits. The team focused on identifying needs, planning and implementing change, and encouraging grass-roots involvement in every phase of the process. One year later, a follow up study showed improved quality of care, retention of positive changes and higher satisfaction among residents, families and staff as compared to the status quo at the project’s onset [ 7 ].
The mini-research groups, an outgrowth of the project’s large group meetings, continued to operate far beyond the official termination of the project (about 10 years). Each group consisted of practical unit nurses, guided by an academic advisor, and focused on a common unresolved clinical problem, which was raised by the clinical staff. With the help of the academic advisors, the problems were framed in terms of systematic research questions, with the goal of formulating appropriate interventions for challenging issues. Favorable results from one study group encouraged the establishment of additional mini-groups to solve other problems within the psychogeriatric unit’s daily routine. Altogether, about 15 mini-research groups were convened to study a wide range of clinical problems, such as how to use Jacuzzi bathing as a therapeutic tool; how to address loneliness; and how to reduce violence.
In order to illustrate how the insights gained from a mini-research project can serve to promote the understanding of dementia and the improvement of care, several exemplars of successful mini-research projects are presented herein, each with its distinctive incentive, methodology and outcomes.
Examples of the mini-research projects
- The “Violence Group” Reducing violence among geropsychiatric residents:
Violent outbursts by residents are common occurrences in geropsychiatric settings. The study team decided to study what triggers outbursts of violence. What cues in the resident’s behavior might indicate a mood change? How should violence be categorized? Which interventions can be helpful?
The group carried out a literature review, gathered more than 30 observed and reported relevant incidents, documented them on a semi-structured questionnaire which they developed (see Fig. 2 ) and, using qualitative techniques, analyzed the data in relation to the residents’ characteristics, the nature of the violent act, the reactions of others, and which interventions were effective. The study team presented its findings to the greater group and its findings encouraged others to establish additional mini-research groups [ 4 ].
- b. The “Mirrors Group” – The use of mirrors as a therapeutic tool for raising self-awareness:

Sample incident report
An occasional observation reported by a nurse about a resident in the geropsychiatric unit ,who was searching obsessively in front of and behind the mirror - provided the incentive to establish another group to examine the effects of mirrors on persons with dementia. How do persons with dementia relate to their image in the mirror? Is the use of mirrors effective in raising levels of self-awareness, calmness and satisfaction? In order to answer these questions, the mini-research group carried out a simple experiment in which 100 persons with dementia were exposed to mirrors of different sizes. Their reactions were documented and analyzed, showing varied responses to looking in the mirror. Most responses were positive (52%) with increased self-awareness regarding personal care, while others were indifferent (10%), or even angered (12%). A majority of residents appeared to benefit from looking at the mirrors. In some instances, the use of mirrors led to improved communication between residents and professional staff. The results of the study team’s work brought to light a new and inexpensive therapeutic tool for persons with dementia: mirrors [ 8 ].
- c. The “Dolls Project” – The use of dolls as a therapeutic tool to awaken pleasurable affective responses:
The therapeutic use of dolls in dementia, though still controversial, is becoming more prevalent at nursing homes and dementia centers. Supporters say that dolls can lessen distress, improve communication and reduce the need for psychotropic medication. Critics say that dolls are demeaning and infantilize seniors. The Dolls mini-research project was a pioneering attempt to systematically examine the influence of dolls as a sensory stimulus to residents in geropsychiatric units [ 9 ]. Using a simple experimental design, the staff placed a variety of human and animal figures in a central location inside the activity rooms of 5 units. Using a pre-coded form, the staff observed reactions to the presence of the dolls, method of selection, type of contact, verbal and body communication, behavior of family members and others, and the emotional impact of the dolls. While the attention span of the residents to the dolls varied from a few moments to several hours, the data revealed that more than half of the 100 residents appeared to be happy with the dolls. The residents usually selected “their” same doll. Touching or holding the dolls elicited pleasure, reassurance, and comfort, often stimulating nonverbal communication, with the potential for verbal communication and better interaction between residents and staff. Thus, the researchers found that dolls can be used therapeutically to awaken pleasurable affective responses in persons with dementia.
- d. The “Jacuzzi Bath Project” – The use of Jacuzzis as a therapeutic tool to address the needs of specific residents.
The Jacuzzi research group was actually formed in order to solve a space-management problem: The luxurious Jacuzzi room in one of the units was reduced to a storeroom because the staff was concerned that entering the tub or bathing might cause residents to feel anxiety, confusion, or might provoke them to violence. The group decided to study whether the Jacuzzi could be used therapeutically. A review of the literature did not produce any relevant information, although hydrotherapy is widely accepted. The methodology incorporated a series of case study analyses. The unit team was encouraged to identify residents whose specific problems might be ameliorated through use of the Jacuzzi. The staff provided an inviting Jacuzzi experience and later evaluated the impact of the treatment in selected situations: a person with aggressive behavior; two night wanderers; and a woman with severe body pain due to arthritis. All the Jacuzzi baths produced a beneficial effect, and the staff overcame their concerns about possible harm to the residents. Consequently, two additional nursing homes participating in the project decided to place Jacuzzis in their geropsychiatric units.
- e. The “Social Networks Project” – Understanding the interpersonal relationships among residents in a geropsychiatric unit.
Several nurses were interested in examining the potential for establishing social networks among residents with dementia and the possible impact on the residents’ quality of life. The nurses wished to see if altering the social environment could enhance relationships. This project later developed into a research thesis conducted by graduate student Perri Cohen [ 10 ]. The methodology chosen was a semi-structured open questionnaire (see Fig. 3 observation schedule). It included the description of a relationship, the morphology of each tie (dyad, triad, or cluster structure), the psycho-social nature of the tie (aggressive/passive/friendly), the degree of symmetry in engagement, the function of the tie (intimacy/being together/help/control etc.) and the identification of the initiator. The depth of the tie and the relationship of the environment to the tie were also observed. Data analysis incorporated qualitative and quantitative methods. The results showed that 44% of the residents with dementia were involved in a consistent social tie of some kind, most often observed as “being together” in a dyad (80%), or in a “concern and help” relationship (66%). The resident’s background variables did not influence the formation of social ties, and neither did his/her cognitive or physical function. Significantly, most of the ties were developed between two residents with different levels of function. This seemed to allow for reciprocity and for the enhancement of self-esteem for both parties. The study concluded that social skills, preferences and abilities were relatively preserved in residents with dementia even for those in the more advanced stages of the disease, suggesting that staff members can play a more active role in facilitating the social environment of the residents than previously thought. For example, staff members can maintain a resident’s grooming and aesthetic appearance to promote social interaction, and can promote a friendly atmosphere in the unit for the overall well-being of residents.

Obsevation Schedules Socialities
Contributions of the mini-research group
The impact of the mini-research groups was multi-dimensional and relatively long lasting. The four major contributions of the project were:
- Improved quality of care - The mini-research projects had a marked effect on the quality of care in the psychogeriatric units. Staff became more sensitive, attentive and knowledgeable to residents’ potentials and needs. Care became more holistic in the sense of integrating physical, psychological, and social aspects. Nursing interventions tended to become more active, creative, evidence-based, and individualized, compared to the regimented care provided prior to the project.
- Increased family involvement - Due to the active role family members played during the project by providing data and feedback to the staff, they became more involved in the unit, They intensified their participation in unit social activities, became closer with the staff and gained an increased general awareness of the needs and potential of their relative and the staff.
- Improved self-image of nursing personnel - Personnel employed in the geropsychiatric units traditionally perceive themselves and others employed at nursing homes as holding the least desirable positions in the work world. Those with the opportunity to advance usually preferred more prestigious work environments than those found at geriatric centers. As a result, nursing home staff included few nurses with academic or post-basic preparation. The geropsychiatric project brought a positive change to the self-image of staff members employed in units which participated in mini-research groups. Such staff members became the center of professional attention and the envy of their colleagues in other geriatric units. The geropsychiatric nurses reported that they felt stimulated and challenged and were more eager to continue in their place of work, an environment which had become exciting and rewarding. They felt that they had become more independent in their practice and more knowledgeable, individually and as a group. They took pride in their new practice, they often documented their projects in video and presented their experiences in professional conferences. Upon termination of the formal project, group members decided to continue on their own. They established a national geropsychiatric nurses association, published their own professional journal “The Forum,” organized their own annual conferences, and with some modifications, continue to function as a strong specialty group organization to this day.
- The merit of collaboration between practitioners and researchers – The frequent meetings of the mini-research groups provided a model for collaboration between academia and providers that enriched both parties and enhanced mini-research group outcomes. The merit of the collaboration for the practicing nurses seemed most obvious. With guidance by an experienced researcher from academia, staff was introduced to new ways of thinking and developed a research approach to their everyday practice. They learned how to identify problems, focus on goals, review literature, gather data, analyze data, and reach conclusions. An academic advisor, acted as a role model and a facilitator to energize the nurses’ potential individually and as a group.
The merit of such collaboration for the academic research advisors, while less obvious, also warrants favorable comment. The close exposure of the researcher to practicing nurses and to daily life in the clinical field provided him/her with new, enriched and grounded perspectives which assured the relevancy and accuracy of research in relation to the reality experienced by subjects. Through the joint experience of collaboration, the dialogue between practitioners and researchers fostered new ways of thinking, mutual learning and appreciation between clinical practice and academia. The researchers learned to frame and prioritize research questions consistent with the questions’ importance to persons with dementia, staff and family members. They found that “small and simple” research questions were at times more helpful than “complicated and sophisticated” ones. The staff proved that with little guidance, they could become astute and creative partners in collecting and analyzing data generated by observations and other qualitative method techniques, so relevant for the study of dementia, yet so complicated to implement [ 11 ]. Collaborating with service personnel also afforded researchers the satisfaction of witnessing the immediate implementation of their research ideas and recommendations.. The combination of direct input and real-life problems in the field, aided by the experience of practitioners and the knowledge of academia researchers proved to be a happier marriage than the hopeful parties could have imagined during their courtship.
What can we learn today from the experiences of the mini-research groups which operated in the past?, and how can we apply the lessons learned to the future? Effective research and treatment of dementia, and improving the quality of life and promoting social inclusion of persons with dementia have been identified as a global public health priority by the World Health Organization [ 12 ]. Yet conducting research into these matters presents complex ethical and methodological issues [ 13 , 14 ]. For example, while obtaining an Advance Research Directive (ARD) is still considered a valid consent in the first stages of dementia, ethics review committees are often reluctant to permit even qualitative methods studies to be conducted on people in advanced stages of dementia. These and other obstacles hamper progress [ 11 ]. The mini-research model is one way of addressing the numerous ethical and legal requirements which hinder advancement. The mini-research group format provides a pragmatic solution, not only in overcoming procedural barriers, but also in stimulating more research and promoting a greater understanding of persons with dementia. The idea of bringing practitioners and researchers together to study and resolve specific issues which arise in clinical settings has innumerable advantages: The model is simple to administer and overcomes bureaucratic and logistical barriers. Mini-research groups can bring about significant and immediate impacts on the quality of care because they examine and work to resolve “real” problems in specific settings. A diverse research team has a greater likelihood to understand persons with dementia. The diversity within mini-research groups increases the likelihood of finding creative paths forward and furthering the professional growth of participating field practitioners and academic researchers, all to the benefit of persons with dementia.
Prof. Bergman started her project with modest funding, but overflowing personal magnetism, enthusiasm, motivation as well as receptivity by the clinical community. In the ensuing years, long term care facilities have become more overwhelmed with clinical, ethical, legal and financial constraints. If improving the care of persons with dementia is indeed a global goal, achieving progress will require not only sufficient resources and infrastructure, but the selection of effective models for advancing knowledge and implementing best practices. The collaboration of practitioners and researchers in mini-research groups can provide an answer to many of the challenges of addressing the needs of persons with dementia. Yet, to ensure such cooperation on a national level and for long lasting periods, every care policy program should develop and assimilate an appropriate research strategy aimed to increase the knowledge and understanding as well as to ensure the provision of quality care for people with dementia and their family members.
Acknowledgements
This paper is dedicated to the beloved Prof. Rebecca (Beccy) Bergman (1919-2015) whose vision, creativity, leadership and unique personality led to the outstanding achievements of this project. Prof. Bergman, a distinguished international leader in her field, was the first nurse to be awarded the Israel Prize for her life-long contribution to society and the nation.
Availability of data and materials
Authors’ contributions.
The author read and approved the final manuscript.
Authors’ information
Hava Golander, RN, MSN, PhD, is an associate professor of nursing (retired) at the Department of Nursing, Tel Aviv University and a senior member of Herczeg Institute for the Study of Aging and Old Age.
Ethics approval and consent to participate
Consent for publication.
Not applicable- only one author
Competing interests
The author declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 24 April 2024

Spatiotemporally resolved colorectal oncogenesis in mini-colons ex vivo
- L. Francisco Lorenzo-Martín ORCID: orcid.org/0000-0003-4717-9338 1 na1 ,
- Tania Hübscher ORCID: orcid.org/0000-0002-2376-712X 1 na1 ,
- Amber D. Bowler ORCID: orcid.org/0000-0002-9439-2839 2 , 3 ,
- Nicolas Broguiere ORCID: orcid.org/0000-0001-9934-4505 1 ,
- Jakob Langer ORCID: orcid.org/0000-0002-0095-1936 1 ,
- Lucie Tillard 1 ,
- Mikhail Nikolaev ORCID: orcid.org/0000-0001-5955-900X 4 ,
- Freddy Radtke ORCID: orcid.org/0000-0003-4315-4045 2 , 3 &
- Matthias P. Lutolf ORCID: orcid.org/0000-0002-5898-305X 1 , 4
Nature ( 2024 ) Cite this article
11k Accesses
1 Citations
373 Altmetric
Metrics details
- Cancer models
- Gastrointestinal cancer
- Lab-on-a-chip
- Tissue engineering
Three-dimensional organoid culture technologies have revolutionized cancer research by allowing for more realistic and scalable reproductions of both tumour and microenvironmental structures 1 , 2 , 3 . This has enabled better modelling of low-complexity cancer cell behaviours that occur over relatively short periods of time 4 . However, available organoid systems do not capture the intricate evolutionary process of cancer development in terms of tissue architecture, cell diversity, homeostasis and lifespan. As a consequence, oncogenesis and tumour formation studies are not possible in vitro and instead require the extensive use of animal models, which provide limited spatiotemporal resolution of cellular dynamics and come at a considerable cost in terms of resources and animal lives. Here we developed topobiologically complex mini-colons that are able to undergo tumorigenesis ex vivo by integrating microfabrication, optogenetic and tissue engineering approaches. With this system, tumorigenic transformation can be spatiotemporally controlled by directing oncogenic activation through blue-light exposure, and emergent colon tumours can be tracked in real-time at the single-cell resolution for several weeks without breaking the culture. These induced mini-colons display rich intratumoural and intertumoural diversity and recapitulate key pathophysiological hallmarks displayed by colorectal tumours in vivo. By fine-tuning cell-intrinsic and cell-extrinsic parameters, mini-colons can be used to identify tumorigenic determinants and pharmacological opportunities. As a whole, our study paves the way for cancer initiation research outside living organisms.
Similar content being viewed by others

Shell buckling for programmable metafluids

Three million images and morphological profiles of cells treated with matched chemical and genetic perturbations

Collective intelligence: A unifying concept for integrating biology across scales and substrates
Cancer arises through the accumulation of genetic lesions that confer unrestrained cell growth potential. Over the past 70 years, both two-dimensional (2D) and three-dimensional (3D) in vitro culture models have been developed to make simplified, animal-free versions of cancers readily available for research 4 . These models successfully portray and dissect a wide range of relatively simple cancer cell behaviours, such as proliferation, motility, invasiveness, survival, cell–cell and cell–stroma interactions, and drug responses, among others 1 , 2 , 4 . However, modelling more complex processes that involve multiple cell (sub)types and tissue-level organization remains a challenge, as is the case for cancer initiation.
The cellular transition from healthy to cancerous is an intricate evolutionary process that is still largely obscure due to the insufficient topobiological complexity of the available in vitro cell culture systems, which precludes de novo tumour generation and the establishment of pathophysiologically relevant tumorigenic models 5 , 6 . Even the current gold-standard organoid-based 3D models, which are often postulated as a bridge between in vitro and in vivo 1 , 3 , 7 , are too simplified for modelling cancer development ex vivo. This is mostly due to (1) their closed cystic structure instead of an in vivo-like apically open architecture 8 ; (2) their short lifespan that requires breaking up the culture every few days for passaging 9 ; (3) their lack of topobiological stability and consistency owing to their stochastic growth in 3D matrices 8 ; and (4) their inability to generate hybrid tissues composed of healthy and cancer cells in a balanced and integrated manner 10 . Various next-generation approaches such as bioprinting and microfabrication technologies have been recently implemented to partially address some of these issues 11 , 12 ; however, none have been able to fully recreate intratumour and intertumour complexity. Consequently, cancer research is still inevitably bound to animal experimentation, which provides a pathophysiologically relevant setting, but forbids high-resolution and real-time analyses of cellular dynamics during oncogenesis. Moreover, these models are economically and ethically costly. Thus, while there is the widespread consensus that animal use in research should be reduced, replaced and refined (the 3 Rs 13 ), this commitment is severely hindered by the insufficient physiological complexity displayed by classical in vitro systems.
Here we postulated that a 3D system able to solve the existing limitations of in vitro cultures could be engineered by leveraging scaffold-guided organoid morphogenesis and optogenetics. Specifically, we developed miniature colon tissues in which cells could (1) be cultured for long durations (several weeks) without the need for breaking the culture through passaging; (2) reproduce the stem-differentiated cell patterning axis in a stable and anatomically relevant topology; (3) be easily mutated and tracked in a spatiotemporally controlled manner; and (4) create a biomechanically dynamic system that allows for tumour emergence while preserving the integrity of the surrounding healthy tissue. These features permit the development of biologically complex tumours ex vivo, bridging the gap between in vitro and in vivo models by providing a high-resolution system that can be used to dissect the molecular factors orchestrating cancer initiation.
Spatiotemporally regulated tumorigenesis
We focused on colorectal cancer (CRC) as it is one of the most prominent cancer types worldwide and its malignant transformation can be readily engineered genetically 14 , 15 . To first achieve spatiotemporal control of oncogenic DNA recombination, we developed a doxycycline-sensitive blue-light-regulated Cre system (hereafter, OptoCre), which we then introduced into inducible Apc fl/fl Kras LSL-G12D/+ Trp53 fl/fl (AKP) healthy colon organoids (Extended Data Fig. 1a–c ). A fluorescent Cre reporter was also incorporated to track cells that undergo oncogenic recombination (Extended Data Fig. 1b,c ). We initially tested the system in conventional organoid cultures, in which OptoCre efficiently induced recombination in the presence of blue light and doxycycline (Extended Data Fig. 1d,e ). Dosage optimization prevented unwanted activation by coupling high efficiency with low leakiness (~1.6%) (Extended Data Fig. 1d,e ). To confirm successful oncogenic transformation, we removed growth factors (EGF, noggin, R-spondin, WNT3A) from the organoid medium and observed that only cells with an activated OptoCre were able to grow, a well-known hallmark of mutated AKP colon organoids 16 (Extended Data Fig. 1f ). The presence of the expected mutations at the Apc, Kras and Trp53 loci was confirmed by PCR and exome sequencing (see below; Extended Data Fig. 3f,g ).
On the basis of previous evidence that small intestine cells can form stable tube-shaped epithelia through scaffold-guided organoid morphogenesis in microfluidic devices 9 , we next aimed to establish a ‘mini-colon’ constituted by OptoCre-AKP cells. By seeding colon cell suspensions in hydrogel-patterned microfluidic devices, we generated single-layered colonic epithelia spatially arranged into crypt- and lumen-like domains (Extended Data Fig. 2a ). This spatial arrangement recapitulated the spatial distribution found in vivo, with stem and progenitor (SOX9 + ) cells located at the bottom of the crypt domains and more differentiated colonocytes (FABP1 + ) located in the upper crypt and lumen areas 17 , 18 (Extended Data Fig. 2b ). In contrast to conventional colon organoids, the lumen of these mini-colons was readily perfusable with fresh medium, enabling the removal of cell debris and extending their lifespan to several weeks without the need for passaging or tissue disruption (Extended Data Fig. 2a ).
Once the healthy mini-colon system was established, we investigated its potential to capture tumour biology by inducing oncogenic recombination through blue-light illumination (Fig. 1a ). To mimic the scenario found in vivo, we fine-tuned OptoCre activation to mutate only a small number of cells (<0.5% of the total population). Due to the stability and defined topology of the mini-colon, we easily detected the acquisition of AKP mutations at the single-cell level (GFP + cells) and tracked their evolution over time (Extended Data Fig. 2c,d ). This revealed that cell death is one of the earliest responses to oncogenic recombination, as mutated mini-colons displayed higher cell shedding rates compared with the controls (Extended Data Fig. 2e ), with a large fraction of the mutated cells undergoing apoptosis (Supplementary Video 1 ). Nevertheless, some mutated cells escaped apoptosis and, after a quiescent period (24–72 h), started dividing at an accelerated pace (Extended Data Fig. 2d ). In conventional organoid cultures, these fast-proliferating mutated cells did not lead to any overt tissular rearrangements (Fig. 1b ), whereas, in the mini-colon system, they developed neoplastic structures over 5–10 days (Fig. 1b ). Furthermore, these mini-colon neoplasias evolved from polyp-like to full-blown tumours, recapitulating in vivo tumorigenesis (Fig. 1b,c and Supplementary Videos 2 and 3 ).
a , Schematic of the experimental workflow followed to induce tumorigenesis in mini-colons. CC, colonocyte; ISC, intestinal stem cell; TA, transit-amplifying cell. b , Bright-field and fluorescence images of time-course tumorigenesis experiments in conventional organoids and mini-colons. Fluorescence signal indicates oncogenic recombination. Scale bars, 200 μm (left) and 75 μm (right). c , Bright-field and fluorescence close-up images of a mini-colon tumour. The red and green signals correspond to healthy and mutated cells, respectively. Scale bar, 25 μm. d , Immunofluorescence images of a mini-colon tumour showing the presence of CD44 (top, green), FABP1 (top, magenta) and nuclei (bottom). Scale bar, 35 μm. e , Multiplicity of tumours emerged in mini-colons of the indicated genotypes after light-mediated oncogenic induction. Statistical analysis was performed using two-way analysis of variance (ANOVA) with Sidak’s multiple-comparison test; ** P = 0.024 (day 6, AKP), ** P = 0.0021 (day 24, A), *** P < 0.0001 (all other conditions). n = 5, 4, 3 and 10 mini-colons for the control, light-induced A, light-induced AK and light-induced AKP conditions, respectively. Data are mean ± s.e.m.
Source Data
Immunostaining analyses revealed that these tumours stemmed from CD44 high cells—a bona fide marker for cancer stem cells in vivo 19 —at the base of the epithelium (Fig. 1d , Extended Data Fig. 2f and Supplementary Video 4 ). Conversely, the bulk of the tumours was composed of cells with different degrees of differentiation, as revealed by the downregulation and upregulation of CD44 and FABP1, respectively (Fig. 1d and Supplementary Video 5 ). This indicated the existence of intratumour heterogeneity in the mini-colon, resembling the in vivo scenario 20 . Consistent with this, histopathological studies showed that these tumours displayed the histological organization characteristic of tubular adenomas (Extended Data Fig. 3a ). To validate their cancerous nature, we performed transplantation experiments in immunodeficient mice and found that mini-colon-derived cancer cells formed tumours in vivo with undistinguishable efficiency from bona fide tumour-derived cancer cells (Extended Data Fig. 3b,c ). Moreover, their histopathological structure was also comparable to the one displayed by primary tumours developed in the colon of AKP mice (Extended Data Fig. 3d ) and included the presence of locally invasive nodules and areas with adenocarcinoma-like features (Extended Data Fig. 3e ).
We confirmed through PCR and exome sequencing that tumour development in the mini-colon was directly associated with the expected mutations at the Apc, Kras and Trp53 loci (Extended Data Fig. 3f,g ). Consistent with this, using organoid lines with a reduced mutational burden ( Apc fl/fl Kras LSL-G12D/+ (hereafter, AK) and Apc fl/fl (hereafter, A)) produced longer latencies in tumour development in a dosage-dependent manner (Fig. 1e and Extended Data Fig. 3h ), demonstrating that mini-colon tumorigenesis can be modulated by the number of oncogenic driver mutations. Collectively, these data show that the mini-colon system enables spatiotemporally controlled in vitro modelling of CRC tumorigenesis with a considerable degree of topobiological complexity.
Context-dependent tumorigenic plasticity
Careful examination of induced mini-colons revealed consistent morphological differences among tumours according to their initiation site, with prominent dense or cystic internal structures arising from the crypt and the luminal epithelium, respectively (see below; Fig. 2b (top)). As mini-colons comprise different types of cells along the crypt–lumen axis (Extended Data Fig. 2b ), we leveraged the spatial resolution provided by OptoCre to investigate whether the initiating cell niche conditioned the morphological and functional features of nascent tumours. To spatially control AKP mutagenesis, we coupled the mini-colon to a photomask restricting blue-light exposure to specific regions of the colonic epithelium (Fig. 2a ), which provided low off-target recombination rates (around 8.5%) (Fig. 2b,c and Extended Data Fig. 4a ). Here again, dense and cystic tumours developed when crypt and lumen epithelia, respectively, were mutationally targeted by blue light (Fig. 2b ). To confirm that this was associated with the differentiation status of the tumour-initiating cell, we cultured mini-colons in either low- or high-differentiation medium before oncogenic induction to shift the proportions of (un)differentiated cells. Low-differentiation conditions produced mini-colons with thicker epithelia, early tumour development and a reduced fraction of cystic tumours (Fig. 2d and Extended Data Fig. 4b,c ). Conversely, high-differentiation conditions produced mini-colons with thinner epithelia, delayed tumour formation and increased cystic tumour frequency (Fig. 2d and Extended Data Fig. 4b,c ). These results indicate that the different environments of the mini-colon can shape tumour fate.
a , Schematic of the experimental workflow followed to spatiotemporally target tumorigenesis in mini-colons. b , Bright-field images of mini-colons that have undergone untargeted (top), crypt-targeted (middle) and lumen-targeted (bottom) tumorigenesis. Targeted areas are indicated by dashed blue lines. The black and white arrows indicate tumours with compact and cystic morphologies, respectively. Scale bar, 75 μm. c , The oncogenic recombination efficiency in targeted and off-target areas in mini-colons. Statistical analysis was performed using two-tailed t -tests; *** P < 0.0001. n = 6 mini-colons per condition. Each point represents one mini-colon. d , Bright-field images of induced mini-colons cultured in low-differentiation (top, WENRNi) and high-differentiation (bottom, ENR) conditions. The black and white arrows indicate tumours with compact and cystic morphologies, respectively. Scale bar, 75 μm. e , Schematic of the different colon organoid lines generated in this work. f , Bright-field images of the indicated colon organoid lines cultured for 2 days in basal medium. Scale bar, 200 μm. g , Metabolic activity (measured using resazurin) of the indicated colon organoid lines cultured in basal medium for the indicated time. Numerical labelling (1–8) was used to facilitate cell line identification. Statistical analysis was performed using two-way ANOVA with Sidak’s multiple-comparison test; *** P = 0.0004 (control), *** P < 0.0001 (all other conditions). n = 3 cultures for each line. For c and g , data are mean ± s.e.m.
To evaluate the functional repercussions of the tumour-initiating niche, we isolated cancer cells from mini-colons enriched in either crypt- or lumen-derived tumours and established organoid cell lines (termed mini-colon AKP) (Fig. 2e ). As a control, we generated AKP mutant organoids by shining blue light onto inducible organoids and kept these mutants in parallel with their mini-colon equivalents, doing the required passages on confluency (termed organoid AKP) (Fig. 2e ). We also established organoid cultures from AKP colon tumours extracted from tamoxifen-treated Cdx2-cre ERT2 AKP mice (termed in vivo AKP) (Fig. 2e ). Notably, in contrast to mini-colons, none of these three types of mutant AKP lines were morphologically distinguishable from healthy non-mutated cells when cultured as organoids (Fig. 1b and Extended Data Fig. 4d ). When we cultured these organoids in basal medium depleted of growth factors (BM; Methods ), both in vivo and crypt tumour-derived mini-colon AKP organoids preserved their proliferative potential (Fig. 2f,g ). Conversely, organoid and lumen tumour-enriched mini-colon AKP lines displayed significantly reduced proliferation rates (Fig. 2f,g ). This was not due to intrinsic cycling defects in any of the organoid lines tested, as these differences were not observed in standard cancer organoid medium (BMGF; Methods and Extended Data Fig. 4e ). As expected, healthy organoids did not grow in any of these conditions (Fig. 2f,g and Extended Data Fig. 4e ). Collectively, these results show that there are context-dependent factors aside from the founding AKP mutations that condition the growth potential of AKP cells. They also indicate that the cells derived from mini-colon crypt tumours recapitulate the growth properties of in vivo CRC cells more faithfully than conventional organoids.
To investigate the molecular programs underpinning these observations, we profiled the transcriptome of the different AKP lines using RNA sequencing (RNA-seq). We first characterized the differences between the two AKP lines derived from conventional systems, in vivo and organoid AKP cells, which also had the biggest disparity in growth potential (Fig. 2g ). According to our previous experiments, in vivo AKP cells upregulated many genes involved in canonical cancer pathways and the promotion of cell growth (Extended Data Fig. 4f,g ). Conversely, these cells downregulated genes associated with cell differentiation, patterning and transcriptional regulation (Extended Data Fig. 4f,g ). To evaluate whether mini-colon AKP cells recapitulated this in vivo AKP transcriptional signature, we performed single-sample gene set enrichment analysis (GSEA) across all of the cell lines. Here, most of the mini-colon AKP lines outscored their organoid AKP counterparts, especially those derived from crypt tumours (Extended Data Fig. 4h ). To investigate the transcriptional divergence between crypt- and lumen-enriched mini-colon AKP cells, we compared the lines with the highest (#v, crypt-enriched) and lowest (#i, lumen-enriched) in vivo AKP signature score (Extended Data Fig. 4h ). These analyses revealed that crypt-derived mini-colon AKP cells upregulated genes involved in WNT signalling, stem cell pluripotency, lipid metabolism and other pathways involved in cancer (Extended Data Fig. 4i ). To identify the potential drivers of growth factor independence among these, we searched for overlaps between AKP lines with high growth potential in BM (in vivo AKP, mini-colon AKP #v). We found that the latter overexpressed a collection of genes that is involved in the activation of MAPK cascades, including receptor tyrosine kinases (RTKs), G-protein-coupled receptors and soluble factors (Extended Data Fig. 5a ). We therefore theorized that these cells were engaging a surplus of MAPK signalling that gave them a greater fitness under growth-factor-poor conditions. To validate this idea, we tested their response to a panel of inhibitors, which confirmed that the growth of AKP lines in BM heavily relied on signals from RTKs (Extended Data Fig. 5b,c ; regorafenib), including KIT (Extended Data Fig. 5b,c ; ripretinib) and FGF receptors (Extended Data Fig. 5b,c ; infigratinib). Corroborating this, the ligands for these RTKs (SCF, FGF2) and others involved in colonocyte clonogenicity (IGF1) 21 could enhance the growth of the AKP lines with poor proliferation potential in BM (Extended Data Fig. 5d,e ). Importantly, all of these dependencies were either reduced or not detectable in conventional CRC organoid medium (BMGF) (Extended Data Figs. 4e and 5b,c ). Taken together, these data indicate that the mini-colon is a plastic system in which context-dependent factors can drive different functional features in CRC cells, including the engagement of ancillary RTK signals that boost their growth potential in challenging environments.
Intra- and intertumour heterogeneity
We hypothesized that the diversity observed in tumour morphology and growth potential reflected clonally distinct tumour types being initiated in the mini-colon. To validate this idea, we performed single-cell transcriptomic profiling of tumour-bearing mini-colons incorporating a genetic cell barcoding system 22 to preserve clonal information (Fig. 3a ). On the basis of bona fide transcriptional markers, mini-colons comprised eight major cell types that were segregated into undifferentiated, absorptive and secretory lineages (Fig. 3b ). Undifferentiated ( Krt20 − ) cells included stem ( Lgr 5 + ), actively proliferating ( Mki67 + ) and progenitor ( Sox9 + Cd44 + ) cells (Fig. 3b,c and Extended Data Fig. 6a ). Mature ( Krt20 + ) absorptive colonocytes constituted the largest fraction of the mini-colon, and included bottom, middle and top colonocytes based on zonation markers 23 (such as Aldob , Iqgap2 and Clca4a ) (Fig. 3b,c and Extended Data Fig. 6a ). Mucus-producing goblet cells ( Muc2 + ) and hormone-releasing enteroendocrine cells ( Neurod1 + ) constituted the secretory compartment (Fig. 3b,c and Extended Data Fig. 6a ). Collectively, this diverse in vivo-like cell composition indicates that mini-colons provide a physiologically relevant context for conducting oncogenesis studies.
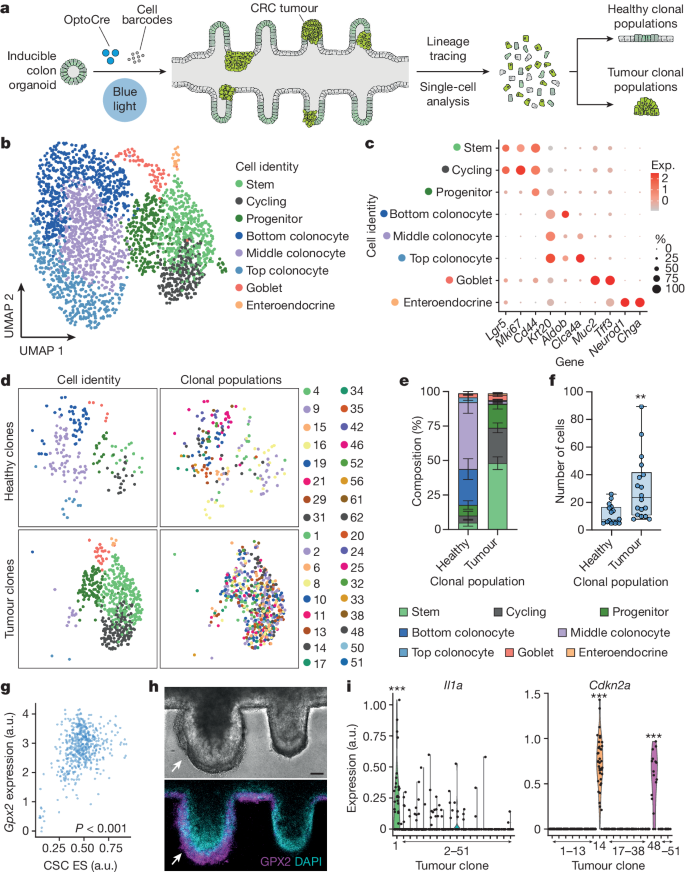
a , Schematic of the experimental workflow followed for single-cell and lineage-tracing analysis of mini-colons. b , Unsupervised uniform manifold approximation and projection (UMAP) clustering of the main cell types in mini-colons 7 days after tumorigenic induction. c , The expression (Exp.) of representative cell-type-specific markers in the different cell populations comprising mini-colons. d , Unsupervised clustering (UMAP) of healthy (top) and tumour (bottom) clonal populations in mini-colons. The cell type (left; colour coded as in b ) and clonal identity (right) are indicated. e , The relative cell type abundance in healthy and tumour mini-colon clonal populations. Data are mean ± s.e.m. n = 16 and 18 for healthy and tumour clones, respectively. f , Healthy and tumour mini-colon clonal population sizes. Statistical analysis was performed using two-tailed Mann–Whitney U -tests; ** P = 0.0011. n = 16 and 18 for healthy and tumour clones, respectively. The box plots show the median (centre lines), the first and third quartiles (box limits) and the minimum and maximum values (whiskers). Each point represents one clonal population. g , The correlation between Gpx2 expression and cancer stem cell transcriptional signature enrichment ( Cd44 , Lgr5 , Sox9 ). Statistical analysis was performed using two-sided Pearson correlation tests; P < 0.0001. n = 540 cells. Each point represents one cell. CSC, cancer stem cell; ES, enrichment score. h , Bright-field and immunofluorescence images showing the abundance of GPX2 (magenta) and nuclei (cyan) in healthy (right) and tumour (left, indicated by arrows) crypts in a mini-colon. Scale bar, 35 μm. i , Expression of the indicated genes in the indicated tumour clones. Statistical analysis was performed using two-sided Wilcoxon rank-sum tests; *** P = 1.77 × 10 −17 ( Il1a , clone 1), 1.00 × 10 −78 ( Cdkn2a , clone 14), 3.67 × 10 −22 ( Cdkn2a , clone 48). n = 540 cells. Each point represents one cell.
To determine the clonal identities across the mini-colon, we compared the genetic barcodes among cells and detected 83 clonal populations. We then discarded small (<5 cells) clones and identified cells containing reads corresponding to the mutated versions of Apc and Trp53 (Extended Data Fig. 6b,c ). These bona fide tumour cells distinguished tumour clonal populations (18 classified) from healthy counterparts (16 classified) ( Methods and Extended Data Fig. 6d ). On average, healthy clonal populations consisted of around 18% undifferentiated cells, which gave rise to the remaining approximately 82% of absorptive colonocytes and secretory cells (Fig. 3d,e ). Conversely, mini-colon tumours were mostly formed by undifferentiated cells (~92%), with sparsely present colonocytes and secretory cells (Fig. 3d,e ). Tumour cells also formed larger clonal populations compared with their healthy counterparts (Fig. 3f ). These cell proportions are well aligned with the ones commonly observed in vivo 24 , 25 .
Analyses of the internal structure of single clonal tumours showed that they comprised a non-homogeneous collection of cells with diverse proliferation, stemness and differentiation markers (Extended Data Fig. 7a ). Such intratumour heterogeneity reflects the complexity of mini-colon tumours, consistent with our immunostaining data (Fig. 1d ). To investigate the mechanisms orchestrating cancer stemness and tumour development, we analysed the transcriptional differences between differentiated ( Krt20 + Apoc2 + Fabp2 + ) and stem ( Lgr5 + Cd44 + Sox9 + ) cancer cells within tumours. We found that Gpx2 , a glutathione peroxidase recently linked to CRC malignant transformation 24 , strongly correlated with the stemness potential of mini-colon cancer cells (Fig. 3g ). Consistent with this, we observed that GPX2 protein was particularly enriched in the basal cells of mini-colon tumours (Fig. 3h ).
To examine whether mini-colons could produce different types of tumours, we next compared the transcriptional profiles of the different tumour clones. Even though all tumour-initiating cells carried the same founding AKP mutations and shared many molecular features, we found clear diversity across mini-colon tumours (Extended Data Fig. 7b ). For example, the expression of the interleukin Il1a and leukocyte peptidase inhibitor Slpi revealed the presence of tumours with an inflammatory-like profile (Fig. 3i and Extended Data Fig. 7b,c ). Cdkn2a (encoding tumour suppressors p14 and p16) and Prdm16 were exclusively expressed by tumours seemingly insensitive to these cell cycle arrest genes given their Ki67 + nature (Fig. 3i and Extended Data Figs. 6a and 7b,c ). Aqp5 , an aquaporin inductor of gastric and colon carcinogenesis 26 , marked specific tumours able to produce the oncogenesis-promoting fibroblast growth factor FGF13 (Extended Data Fig. 7b,c ). Together with other markers (Extended Data Fig. 7b ) and corroborations at the protein level (Extended Data Fig. 7d ), these data indicate that a variety of tumour subtypes can be generated in the mini-colon, arguably due to tumour-niche-intrinsic and/or environmental factors. This probably accounts for the observed differences among mini-colon AKP cell lines (Fig. 2g and Extended Data Fig. 4h ). Importantly, we found that this diversity was relatable to the human context. For example, mini-colons generated tumours with transcriptional profiles representing both iCMS2- and iCMS3-like subtypes 27 (Extended Data Fig. 8a,b ) that were associated with a wide range of aggressiveness profiles (Extended Data Fig. 8c ) and correlated with different extents of lymph node colonization (Extended Data Fig. 8d,e ) when cross-compared with transcriptomic data from the TCGA collection of patients with CRC. Collectively, these findings demonstrate that the mini-colon is a complex cellular ecosystem that recreates both healthy and cancer cell diversity.
Screening of tumorigenic factors
The longevity, experimental flexibility and tumour formation dynamics of mini-colons provides an unparalleled in vitro set-up for conducting tumorigenesis assays. We therefore next used mini-colons as screening tools for identifying molecules with a prominent role in tumour development. As our single-cell RNA-seq (scRNA-seq) analyses revealed Gpx2 overexpression in cancer stem cells (Fig. 3g,h ), we probed its functional relevance by adding the glutathione peroxidase inhibitor tiopronin 28 to the basal medium reservoirs of mini-colons right after blue-light-induced AKP mutagenesis (Fig. 4a ). Basal application of the drug provides ubiquitous exposure on the mini-colon basolateral domain, mimicking a systemic therapy model (Fig. 4a ). By the time control mini-colons developed full-blown tumours, tiopronin-treated counterparts were largely tumour-free with a healthy colonic epithelium (Fig. 4b and Extended Data Fig. 9a ). This was not due to the mere reduction in proliferative activity, as tiopronin had a minor impact on organoid growth (Extended Data Fig. 9b,c ). As tiopronin targets several glutathione peroxidases, we corroborated the specific implication of GPX2 in tumour initiation by knocking down its transcript (Extended Data Fig. 9d ). These knockdown cells showed no detectable defects in terms of organoid morphology or proliferation in unchallenged conditions (Extended Data Fig. 9e,f ). However, after blue-light-mediated oncogenic recombination, GPX2-deficient mini-colons developed tumours with reduced kinetics and multiplicity (Fig. 4c and Extended Data Fig. 9g ), recapitulating the results obtained with tiopronin (Fig. 4b and Extended Data Fig. 9a ). Importantly, mini-colons were instrumental for these findings, as conventional organoid cultures cannot reveal differences in tumour-forming abilities (Extended Data Fig. 9b,h ).
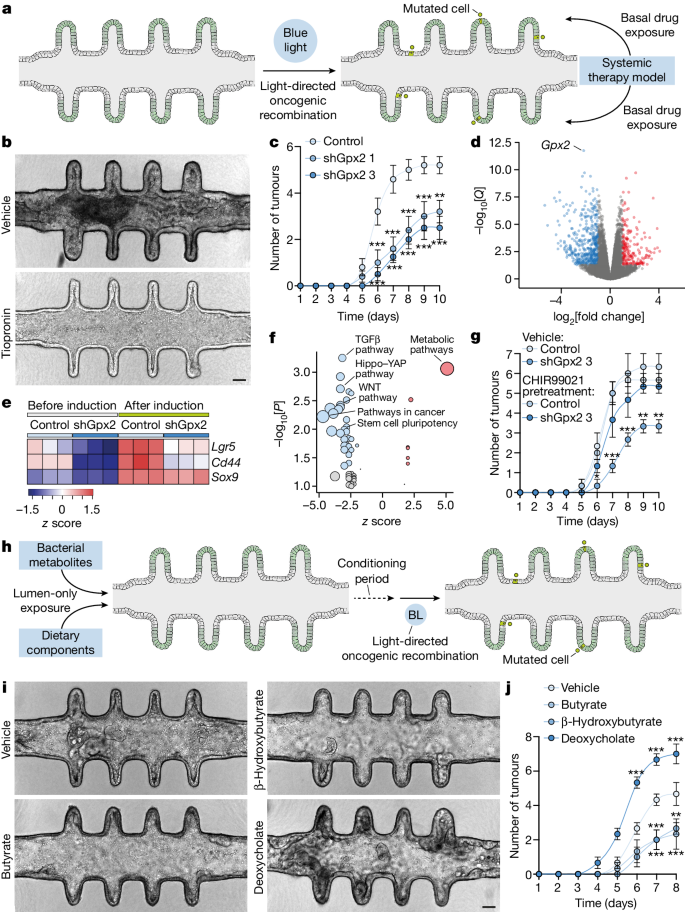
a , The experimental workflow for systemic therapy modelling. b , Bright-field images of mini-colons treated with vehicle or tiopronin after tumorigenic recombination. Images correspond to 6 days after induction. Scale bar, 75 μm. c , The multiplicity of tumours emerged in mini-colons of the indicated genotype after oncogenic induction. Statistical analysis was performed using two-way ANOVA with Sidak’s multiple-comparison test; ** P = 0.0034, *** P = 0.0007 (days 6 and 9, sh Gpx2 1), *** P < 0.0001 (all other conditions). n = 5, 5 and 4 mini-colons for control, shGpx2 1 and shGpx2 3, respectively. d , Differentially expressed genes after Gpx2 knockdown in light-induced AKP tumour cells. e , Expression of the indicated genes in colonocytes of the indicated genotypes before and after oncogenic recombination. The colour scale shows the z score. f , The main enriched functional terms after Gpx2 knockdown in light-induced AKP tumour cells. Significant terms are highlighted in blue or red, as determined using one-sided Fisher’s exact tests, with gene expression adjusted P -values (Benjamini-Hochberg correction). g , The multiplicity of tumours emerged in mini-colons of the indicated genotype under the indicated pretreatment (2 days before oncogenic induction). Statistical analysis was performed using two-way ANOVA with Sidak’s multiple-comparison test; * P = 0.0274, ** P = 0.0033 (days 9 and 10), *** P < 0.0001 (days 7 and 8). n = 3 mini-colons for each condition. h , The experimental workflow for microbiota and dietary pattern modelling. BL, blue light. i , Bright-field images of mini-colons treated with the indicated metabolites. Images correspond to 7 days after tumorigenic induction. Scale bar, 75 μm. j , The multiplicity of tumours emerged in mini-colons treated with the indicated metabolites. Statistical analysis was performed using two-way ANOVA with Sidak’s multiple-comparison test; ** P = 0.0080, *** P = 0.0008 (days 7 and 8), *** P < 0.0001 (day 6). n = 3 mini-colons for each condition. For c , g and j , data are mean ± s.e.m.
To gain molecular insights into the mechanism engaged by GPX2, we performed RNA-seq analysis of Gpx2 -knockdown cells both before and after oncogenic recombination. These analyses revealed that GPX2 deficiency remodels the colonocyte transcriptome in both healthy (Extended Data Fig. 9i ) and tumorigenic (Fig. 4d ) conditions (Supplementary Tables 1 and 2 ). This included the downmodulation of canonical markers associated with both healthy and cancer cell stemness, such as Lgr5 and Cd44 (Fig. 4e ). By contrast, markers of proliferative progenitor cells, such as Sox9 , remained unchanged (Fig. 4e ). Consistent with this, Gpx2 abrogation led to the repression of transcriptional programs implicated in stem cell pluripotency, including the WNT, Hippo–YAP and TGFβ pathways, as well as epithelial–mesenchymal transition and other processes involved in cancer cell fitness (Fig. 4f and Extended Data Fig. 9j,k ). Conversely, transcriptional programs associated with proliferation were not affected, consistent with our observations in cell culture (Extended Data Fig. 9e,f,k ). These findings indicate that the inhibition of GPX2 downmodulates colonocyte stemness, which probably accounts for the reduced tumorigenic potential observed in the mini-colon after oncogenic recombination. Supporting this, we found that non-transformed GPX2-deficient cells displayed reduced clonogenic capacity in medium deprived of exogenous WNT signals (Extended Data Fig. 9l,m ). Furthermore, the enhancement of WNT signalling through pretreatment of mini-colons with CHIR99021 for 2 days before oncogenic induction rescued the tumorigenic potential of Gpx2- knockdown cells (Fig. 4g and Extended Data Fig. 9n ). Collectively, these data uncover GPX2 as a key regulator of colon stemness and tumorigenesis, shedding light on lingering questions spurred by the recent discovery of its association with the malignant progression of human CRC 24 .
Besides cell-intrinsic factors, colon tumorigenesis in vivo is heavily modulated by a myriad of environmental molecules that continuously contact the luminal side of colonocytes, such as the metabolites produced by colon-residing microbiota 29 . The impact of these molecules cannot be faithfully evaluated in conventional organoid cultures, as their lumen is not accessible. As mini-colons address this limitation, we also investigated whether they could model the role of bacterial metabolites of which the tumorigenic function has been corroborated in vivo. To that end, we administered specific metabolites exclusively in the luminal side of healthy mini-colons and, after a conditioning period of 2 days, induced oncogenic recombination (Fig. 4h ). When luminally exposed to deoxycholic acid, a tumour-promoting metabolite 29 , 30 , 31 , mini-colons developed tumours with fast kinetics and high multiplicity (Fig. 4i,j ). Conversely, both tumour-suppressive butyrate 29 , 32 and β-hydroxybutyrate 33 slowed tumour development and reduced multiplicity (Fig. 4i,j ). These results demonstrate that mini-colons faithfully recapitulate the in vivo pathophysiological responses to bacterial metabolites, whereas conventional organoid cultures do not provide informative data on their tumorigenic relevance (Extended Data Fig. 10a ).
Dietary components also constitute a relevant source of luminal factors conditioning colon tumorigenesis 34 . We therefore performed analogous experiments modelling diets with different caloric contents (Fig. 4h and Extended Data Fig. 10b ). These revealed that calorie restriction in the luminal space effectively reduced tumour burden when compared to calorie-enriched medium (Extended Data Fig. 10c,d ), consistent with in vivo evidence 35 . To show the relevance of luminal accessibility, we placed the same amount of dietary medium in the basal medium reservoirs instead of the luminal space (Extended Data Fig. 10b ). Here, no differences were observed between the two dietary patterns (Extended Data Fig. 10e,f ), therefore indicating that an accessible lumen—a forbidden feature in conventional organoids—is decisive for the physiologically relevant modelling of colon biology. Collectively, these findings demonstrate that the mini-colon is a versatile tool that enables faithful in vitro recapitulation of CRC tumorigenesis and its environmental determinants.
Here we show that the mini-colon model shifts the paradigm of cancer initiation research, allowing ex vivo tumorigenesis with unparalleled pathophysiological intricacy. Coupled with spatiotemporal control of oncogenesis, real-time single-cell resolution and broad experimental flexibility, this system opens new perspectives for in vitro screening of cellular and molecular determinants of cancer development. Supporting this, mini-colons faithfully reflect in vivo-like responses to microbiota-derived metabolites and dietary patterns. Likewise, our model can help in the discovery and validation of genetic targets and tumour-suppressive drugs, as illustrated by the finding that glutathione peroxidase inhibition abrogates CRC tumour development. This constitutes a major advance over conventional 3D culture systems like organoids and Transwell models, which can recapitulate isolated aspects of colon biology such as histopathological features 36 or apical accessibility 37 , respectively, but lack the all-round topobiological complexity required to allow tumour formation ex vivo. Although such complexity demands bioengineering expertise to generate mini-colons, we have provided a detailed protocol that makes this system widely adoptable across laboratories that are already familiar with conventional organoid cultures (Protocol Exchange 38 ; see Methods ).
As for most genetic models of CRC, our system is based on the simultaneous acquisition of several mutations, which does not fully recapitulate the sequential tumorigenic process that occurs in vivo 39 . We therefore acknowledge that adopting a stepwise mutational system will enhance the relevance of the mini-colon as a cancer initiation model. We are also aware that spatial transcriptomics approaches will improve our understanding of tumour heterogeneity in the mini-colon. In the same lines, we envision the incorporation of additional regulatory layers in our OptoCre system, such as the fusion with the oestrogen receptor ligand-binding domain for subcellular localization control 40 , as a promising way to achieve finer spatiotemporal regulation of recombination.
Although mini-colons cannot be considered to be a general replacement for animals in all contexts of cancer research, they offer the possibility to reduce animal use in a wide range of experimental applications. Importantly, the pathophysiological relevance of the mini-colon can be readily enhanced by including stromal cells in the surrounding biomimetic extracellular matrix, which condition both tumour dynamics and invasiveness (Extended Data Fig. 10g–k ). Current lines of work that will be made available in ensuing publications have also proved that this model can be applied to patient-derived colorectal cancer specimens. Lastly, we anticipate that, by adapting its biomechanical properties, topology and culture conditions, it will be possible to expand the system to other prominent epithelial cancer types, such as lung, breast or prostate, bringing an important experimental resource to multiple fields.
Apc fl/fl mice (a gift from T. Petrova) were crossed to Cdx2-cre ERT2 mice (The Jackson Laboratory). Apc fl/fl Cdx2-cre ERT2 mice (termed A) were then crossed with Kras LSL-G12D/+ Trp53 fl/fl mice (a gift from E. Meylan) to generate Apc fl/fl Kras LSL-G12D/+ Trp53 fl/fl Cdx2-cre ERT2 mice (termed AKP). AKP mice were then back-crossed with C57BL6/J (The Jackson Laboratory) to generate Apc fl/fl Kras LSL-G12D/+ Cdx2-cre ERT2 mice (termed AK).
To induce tumorigenesis in vivo, Cre ERT2 recombinase was activated at the age of 8–10 weeks by a single intraperitoneal injection of 18 mg kg –1 tamoxifen (Sigma-Aldrich, T5648) in sunflower oil. Tumours were allowed to develop for 6 weeks. Mice were then sacrificed for tissue and cell isolation. See also below the specific section for transplantation of organoids in immunocompromised mice.
All animal work was conducted in accordance with Swiss national guidelines, reviewed and approved by the Service Veterinaire Cantonal of Etat de Vaud (VD3035.1 and VD3823). These regulations established 800 mm 3 as the maximal subcutaneous tumour volume allowed, which was not exceeded in any of the experiments. In experiments in which tumorigenesis was induced in vivo, the locomotion, appearance, body condition and intestinal function of the mice were monitored twice weekly and assigned numerical scores to allow quantitative decision making in case humane end points were necessary before the predefined end point of the experiment (6 weeks). All of the mice in this study reached the predefined end point. Mice were kept in the animal facility under EPFL animal care regulations. They were housed in individual cages at 23 ± 1 °C and 55 ± 10% humidity under a 12 h–12 h light–dark cycle. All of the animals were supplied with food and water ad libitum.
OptoCre module plasmid generation
The OptoCre module was designed by integrating the following constructs: (1) FUW-M2rtTA, which constitutively expresses the reverse tetracycline transactivator (rtTA); (2) FUW-tetO-GAVPO, which expresses the light-switchable trans-activator GAVPO after rtTA binding in the presence of doxycycline; and (3) FUW-OptoCre, which expresses Cre recombinase after GAVPO binding in the presence of blue light (Extended Data Fig. 1a ). FUW-M2rtTA was purchased from Addgene (20342). Vectors containing GAVPO and the GAVPO-binding promoter (UASG) 5 -P min , developed previously 41 , were a gift from M. Thomson 42 . For FUW-tetO-GAVPO generation, GAVPO was subcloned into the doxycycline-responsive FUW-TetO backbone (Wernig Lab, Stanford) using the EcoRI and NheI restriction sites (Extended Data Fig. 1a ). For FUW-OptoCre generation, (UASG) 5 -P min was inserted into the FUW-TetO backbone from which the TetO promoter had been removed (Wernig Lab, Stanford) using the BstBI and BamHI restriction sites. We then introduced the Cre recombinase (Addgene, 25997) downstream of (UASG) 5 -P min using the Pac1 restriction sites (Extended Data Fig. 1a ).
Isolation of colon cells
Healthy colon or tumour pieces were finely chopped using a scalpel and transferred to a gentle-MACS C-tube (Miltenyi, 130-093-237) containing 4 ml of digestion medium (RPMI (Thermo Fisher Scientific, 22400089), 1 mg ml –1 collagenase type IV (Life Technologies, 9001-12-1), 0.5 mg ml –1 dispase II (Life Technologies, 17105041) and 10 μg ml –1 DNase I (Applichem, A3778)). Tissues were then digested using the 37C_m_TDK_1 program on the gentle-MACS Octo Dissociator with heaters (Miltenyi). After the program was complete, the cell suspension was passed through a 70-μm strainer (Corning, 431751) and centrifugated at 400 g for 5 min.
Organoid and stromal cell culture
To establish organoids, colon cells were embedded in growth-factor-reduced Matrigel (Corning, 356231) (~2 × 10 4 cells per 20 μl dome) and cultured in Advanced DMEM/F-12 (Thermo Fisher Scientific, 12634028) supplemented with 1× GlutaMax (Thermo Fisher Scientific, 35050038), 10 mM HEPES (Thermo Fisher Scientific, 15630056), 100 μg ml −1 penicillin–streptomycin (Thermo Fisher Scientific, 15140122), 1× B-27 supplement (Thermo Fisher Scientific, 17504001), 1× N2 supplement (Thermo Fisher Scientific, 17502001), 1 mM N -acetylcysteine (Sigma-Aldrich, A9165), 50 μg ml −1 primocin (InvivoGen, ant-pm-2), 50 ng ml −1 EGF (Peprotech, 315-09), 100 ng ml −1 noggin (produced at EPFL Protein Production and Structure Core Facility), 500 ng ml −1 R-spondin (produced at EPFL Protein Production and Structure Core Facility), 50 ng ml –1 WNT3A (Time Bioscience, rmW3aL-010), 10 mM nicotinamide (Calbiochem, 481907) and 2.5 μM Thiazovivin (Stemgen, AMS.04-0017). This full medium is termed ‘WENRNi’. The base version of this medium without EGF, noggin, R-spondin, WNT3A and nicotinamide is referred to as BMGF and was used for the expansion of colon tumour organoids since they do not require the additional growth factors. The base version of BMGF without B-27, N2 and N -acetylcysteine is termed BM or basal medium, and was used for growth-factor deprivation experiments. A detailed protocol describing organoid culture can be found elsewhere 9 . Where indicated, organoids were treated with the following compounds or growth factors: regorafenib (8 μM, Selleckchem, S1178), ripretinib (1 μM, Selleckchem, S8757), infigratinib (1 μM, Selleckchem, S2183), SCF (100 ng ml –1 , PeproTech, 250-03), FGF2 (50 ng ml –1 , Thermo Fisher Scientific, PMG0035) and IGF1 (100 ng ml –1 , R&D Systems, 291-G1-200). Stromal cells were derived from cell suspensions from the primary tissue cultured in EGM-2 MV Microvascular Endothelial Cell Growth Medium-2 (Lonza, CC-3202) on conventional cell culture flasks. This medium selection strategy was followed by magnetic-activated cell sorting (MACS) on EPCAM (Miltenyi Biotec, 130-105-958) according to the manufacturer’s instructions to discard epithelial cells. The presence of stromal cells was further confirmed by immunofluorescence analyses of vimentin expression (see below). Cells were tested for mycoplasma before cryopreservation and in randomized routine checks using the MycoAlert PLUS Mycoplasma Detection Kit (Lonza, LT07-705).
Generation of light-inducible cells
Lentiviral particles carrying the three components of the OptoCre module (see above; Extended Data Fig. 1b ) and a Cre recombination reporter were produced at the EPFL Gene Therapy Platform by transfecting HEK293 cells with each plasmid of the OptoCre module and pLV-CMV-LoxP-DsRed-LoxP-eGFP (Addgene, 65726) plasmids. Lentivirus-containing supernatants were collected and concentrated by centrifugation (1,500 g for 1 h at 4 °C). Lentiviral titration was performed using the p24-antigen ELISA (ZeptoMetrix, 0801111). For transduction, colon organoids (around 2 × 10 5 cells) were dissociated into single cells by incubating in TrypLE Express Enzyme (Thermo Fisher Scientific, 12605028) at 37 °C for 5 min. Cells were then washed with basal medium supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, 10500064) and resuspended in WENRNi medium containing 8 μg ml −1 polybrene (Sigma-Aldrich, TR-1003-G) and the following amounts of viral particles: ~10 ng of p24 FUW-M2rtTA per ml, ~80 ng of p24 FUW-tetO-GAVPO per ml, ~80 ng of p24 FUW-OptoCre per ml and ~1,000 ng of p24 CMV-LoxP-DsRed-LoxP-eGFP per ml. These cells were plated in a 24-well plate, centrifuged at 600 g for 60 min at room temperature, and incubated for 6 h at 37 °C. After incubation, the cells were collected, centrifuged, plated in 20 μl Matrigel domes in a 24-well plate and cultured in WENRNi medium. Cells expressing the Cre recombination reporter were selected by supplementing WENRNi medium with 8 μg ml −1 puromycin (InvivoGen, ant-pr-1).
Light-mediated oncogenic recombination
The OptoCre module requires (1) doxycycline to induce rtTA-mediated GAVPO expression and (2) blue light to induce GAVPO-mediated Cre recombinase expression (Extended Data Fig. 1a,b ). At the desired time of oncogenic induction, 2 μg ml −1 doxycycline hydrochloride (Sigma-Aldrich, D3072) was added to the culture medium of either the organoids or mini-colons. Light induction was then performed using a custom-made LightBox built by Baur SA and the Instant Lab at EPFL. The LightBox consisted of an Acqua A5 System (Acme Systems) that could be remotely parametrized using a custom-made web-based application. Communication between the Acqua A5 System and the microcontroller (PJRC, Teensy 3.2) was done through Blocky programming, which allowed for control of the LED drivers (Sparkfun, PicoDuck). The LEDs (Cree LEDs, XLamp XP-C Blue LEDs) were placed into a custom multilayer 24-well plate holder made of black anodized aluminium and polyphenylsulfone; the height was optimized for homogeneous light distribution within each well. The entire LightBox, plate-holder, LEDs and cables were made to be placed in the incubator (watertight and heat resistant). Diffusive elements (Luminit, Light Shaping Diffuser 80°) were used to render the illumination more homogeneous inside each well. The intensity of the blue light (450–465 nm, peak at 455 nm) was optimized, set to 100 μW cm −2 and shined on the cells for 3 h. After blue-light exposure, doxycycline was removed by washing the cultures with fresh medium. In experiments targeting the light to specific regions of the mini-colon, work was carried out in the dark using a near infrared light (Therabulb, NIR-A) to prevent leaky Cre expression. Light-targeting was performed using a photomask that was adapted to the dimensions of the mini-colon and that was created from a photoresist and chrome-coated standard 5 × 5 inch silica plate (Nanofilm) with an automated machine (VPG200 Heidelberg Instrument, 2.0 µm resolution). Once the exposed photoresist was developed, the chrome layer was wet-etched and the remaining photoresist was stripped using a mask processor (Hamatech HMR900) 9 .
Microdevice design, fabrication and loading
The microfluidic device used for mini-colon cultures was designed using Clewin 3.1 (Phoenix Software) and fabricated as previously described 9 . It was composed of three main compartments: (1) a hydrogel chamber for cell growth in the centre; (2) two basal medium reservoirs flanking the hydrogel compartment; and (3) inlet and outlet channels for luminal perfusion 9 . An extracellular matrix containing 80% (v/v) type I collagen (5 mg ml −1 , Reprocell, KKN-IAC-50) and 20% (v/v) growth–factor-reduced Matrigel was loaded into the hydrogel compartment. The microchannels constituting the mini-colon architecture within the hydrogel were designed using Adobe Illustrator CC 2019 and Wolfram Mathematica 11.3. They were then read by PALM RoboSoftware 4.6 (Zeiss) and ablated using a nanosecond laser system (1 ns pulses, 100 Hz frequency, 355 nm; PALM Micro-Beam laser microdissection system, Zeiss). The dimensions of the mini-colon architecture were described previously 9 . A detailed description of all the key steps required for the generation and maintenance of mini-guts is available at Protocol Exchange ( https://doi.org/10.21203/rs.3.pex-903/v1 ) 38 .
Mini-colon culture, development and tumorigenesis
Colon organoids were dissociated into single cells by incubating in TrypLE Express Enzyme for 5 min at 37 °C followed by vigorous pipetting. This cell suspension was washed in 5 volumes of Advanced DMEM/F-12 supplemented with 10% FBS and passed through 40 μm cell strainers (Corning, 431750). After centrifugation at 400 g for 5 min, cells were resuspended in WENRNi medium at around 10 6 cells per ml. The mini-colon luminal microchannel was filled with 10 μl of this cell suspension. Cells were allowed to settle down in the mini-colon crypt-shaped cavities for 5 min, and the leftover unadhered cells were washed out from the microchannel by medium perfusion. The basal medium reservoirs were filled with 100 μl of WENRNi. Unless otherwise indicated, once the healthy colonic epithelium was fully formed (around 2 days after seeding), the medium in the luminal channel was switched to BM, while WENRNi was kept in the basal medium reservoirs. This gradient of growth factor from basal medium reservoirs to luminal space favours colonocyte differentiation across the crypt–lumen axis. For low-differentiation conditions of the differentiation experiments, WENRNi was kept in both the lumen and basal medium reservoirs. Conversely, high-differentiation mini-colons were cultured in WENRNi medium without WNT3A and nicotinamide (termed ENR). Unless otherwise stated, once the colonic epithelium was fully formed, oncogenic induction in the mini-colons was performed as stated above. Where indicated, tiopronin (5 mM, Selleckchem, S2062) or CHIR99021 (3 μM, StemCell Technologies, 100-1042) was added to the basal medium reservoirs after or before oncogenic induction, respectively. For co-culture experiments, ~500 stromal cells were seeded in each hydrogel before the laser-mediated ablation of the mini-colon pattern. The rest of the culture conditions and procedures remained unchanged. To avoid potential unspecific results derived from the small (but non-zero; Extended Data Fig. 1d,e ) leakiness of the optogenetic system, each replication across all studies was performed using independent OptoCre organoid lines freshly generated before each experiment. In all cases, the mini-colons were incubated at 37 °C in 5% CO 2 humidified air, with daily luminal perfusions and medium changes every other day.
Mini-colon whole-mount immunofluorescence staining
Mini-colons were rinsed with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde (Thermo Fisher Scientific, 15434389) overnight at 4 °C. After rinsing with PBS, the hydrogels were extracted from the PDMS scaffold using a scalpel, placed into a 48-well plate, permeabilized with 0.1% Tween-20 (Sigma-Aldrich, P9416) in PBS (10 min at 4 °C) and blocked in 2 mg ml −1 bovine serum albumin (Sigma-Aldrich, A3059) in PBS containing 0.1% Triton X-100 (Sigma-Aldrich, T8787) (blocking buffer) for at least 45 min at 4 °C. The samples were subsequently incubated overnight at 4 °C in blocking buffer with the corresponding following primary antibodies: CD44 (1:200; Abcam, ab157107), FABP1 (1:100; R&D Systems, AF1565), SOX9 (1:200; Abcam, ab185966), GPX2 (1:200; Bioss Antibodies, BS-13396R), IL-1α (1:200; R&D Systems, AF-400-SP), CDKN2A (1:100; Abcam, ab211542), E-cadherin (1:100; Abcam, ab11512) and vimentin (1:200; Abcam, ab92547). After three washes in blocking buffer for a total of 6 h at room temperature, the samples were incubated overnight at 4 °C in blocking buffer with the following corresponding secondary antibodies: Alexa Fluor 488 anti-goat (1:400, Thermo Fisher Scientific, A-11055), Alexa Fluor 488 anti-rat (1:400, Thermo Fisher Scientific, A-21208) and Alexa Fluor 647 anti-rabbit (1:400, Thermo Fisher Scientific, A-31573). After 3 washes in blocking buffer for a total of 6 h at room temperature, the samples were incubated with DAPI (1 μg ml −1 ; Tocris Bioscience, 5748) for 10 min at room temperature in blocking buffer. Before imaging, the hydrogels were mounted onto 35 mm glass bottom dishes (Ibidi, 81218-200) in Fluoromount-G (SouthernBiotech, 0100-01).
Mini-colon sectioning and histochemistry
Mini-colons were fixed and extracted from the PDMS scaffold as indicated above and were prepared for cryosectioning by incubating in 30% (w/v) sucrose (Sigma-Aldrich, S1888) in PBS until the sample sank. Subsequently, the samples were incubated for 12 h in a mixture of Cryomatrix (Epredia, 6769006) and 30% sucrose (mixing ratio 50/50) followed by a 12 h incubation in pure Cryomatrix. The samples were then embedded in a tissue mould, frozen on dry ice, and cut into 40-µm-thick sections at −20 °C using the CM3050S cryostat (Leica). Haematoxylin and eosin staining was performed at the EPFL Histology Core Facility using the Ventana Discovery Ultra automated slide preparation system (Roche).
Microscopy and image analysis
Bright-field and fluorescence imaging of living organoids and mini-colons was performed using the Nikon Eclipse Ti2 inverted microscope with ×4/0.13 NA, ×10/0.30 NA and ×40/0.3 NA air objectives and a DS-Qi2 camera (Nikon Corporation). Time lapses were taken in a Nikon Eclipse Ti inverted microscope system equipped with ×4/0.20 NA and ×10/0.30 NA air objectives and DS-Qi2 (Nikon Corporation) and Andor iXon Ultra 888 (Oxford Instruments) cameras. Both systems were controlled using the NIS-Elements AR software (Nikon Corporation). The extended depth of field (EDF) of bright-field images was calculated using a built-in NIS-Elements function. Fluorescence confocal imaging of fixed mini-colons was performed using the Leica SP8 STED 3X inverted microscope system equipped with ×10/0.30 NA air and ×25/0.95 NA water objectives, 405 nm diode and supercontinuum 470–670 nm lasers, and the system was controlled by the Leica LAS-X software (v.3.5.7, Leica microsystems). Histological sections were imaged using a Leica DM5500 upright microscope with ×10/0.30 NA and ×20/0.75 NA air objectives, a ×40/1.0 NA oil objective and a DMC 2900 Color camera, and the system was controlled by the Leica LAS-X software. Image processing was performed using standard contrast- and intensity-level adjustments in ImageJ (NIH). For oncogenic recombination analyses, the GFP-positive area was measured from 16-bit EDF images by subtracting the background, sharpening the images, and applying a signal threshold and a mask. The ratio between GFP-positive area and total organoid area was used for analyses. Recombined cells were segmented using StarDist with the default parameters ( https://github.com/stardist ) on the GFP channel of mini-colon images. Cell debris was discarded from segmentation analyses by setting an empirically established size threshold. For tumour quantification in the mini-colon, neoplastic structures with at least three times the thickness of the surrounding healthy epithelium were considered to be tumours. Videos of immunostainings were rendered using Imaris (Oxford Instruments).
Mini-colon shedding evaluation
The medium from the luminal compartments of the mini-colons, together with an additional luminal perfusion of 10 μl of basal medium, was collected every day for 4 days after the blue-light-induced oncogenic recombination. The protein content in these extracts was analysed using conventional Bradford assays (Bio-Rad, 5000006) and used as an indicator of cell shedding.
Mini-colon cell line derivation
Mini-colon-containing hydrogels were extracted from their microfluidic devices with a scalpel as indicated above and incubated with 0.1% (w/v) collagenase I (Thermo Fisher Scientific, 17100-017) at 37 °C for 10 min. Once the hydrogel was fully digested, the mini-colon was washed with PBS and digested with TrypLE Express Enzyme for 5 min at 37 °C. The resulting cell suspension was washed with Advanced DMEM/F-12 supplemented with 10% FBS, pelleted, embedded in Matrigel and cultured as indicated above for regular colon organoids.
Transplantation of organoids in immunocompromised mice
Organoid lines were established as indicated above from either in vivo colon tumours (reference AKP) or tumour-bearing mini-colons (mini-colon AKP). These organoids were dissociated into single cells using TrypLE Express Enzyme for 5 min at 37 °C, washed with Advanced DMEM/F-12 supplemented with 10% FBS, pelleted and embedded in Matrigel at 2.5 × 10 6 cells per ml. A total of 100 μl of this suspension was inoculated by subcutaneous injection into the right flank of NOD. Cd-Prkdz scid Il2rg tm1Wjl /Szj (NSG) mice (Jackson laboratories). Tumour growth was monitored using callipers twice per week until the end point at 18 days after inoculation. Length ( L ) and width ( W ) were measured and used to approximate the volume ( V ) of the tumour in mm 3 using the modified ellipsoid formula: V = ( L × W 2 )/2. After euthanasia, tumours were resected from the graft location and measured once more with callipers.
Graft sectioning and histochemistry
Tumour samples were fixed overnight in 4% paraformaldehyde at 4 °C, dehydrated in graded ethanol baths, cleared with xylene, embedded in paraffin and cut into 4-µm-thick sections using the HM 325 Rotary Microtome (Thermo Fisher Scientific). These sections were mounted onto Superfrost plus slides (Epredia, J1800AMNZ) and allowed to dry for 2 days at room temperature. Haematoxylin and eosin staining was performed at the EPFL Histology Core Facility using the Ventana Discovery Ultra automated slide preparation system (Roche).
Mutational screening in colon organoids
Genomic DNA was isolated from colon cells using the PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific, K182001) according to the manufacturer’s instructions. Recombination of the LSL (LoxP-Stop-LoxP) cassette controlling Kras G12D expression was confirmed by PCR using the protocol and oligos described by the Tyler Jacks laboratory ( https://jacks-lab.mit.edu/ , Kras G12D Conditional PCR). Apc and Trp53 recombinations were confirmed through exome sequencing performed at BGI Genomics at 100× coverage using DNBSEQ sequencing technology. DNA reads were mapped to the mouse GRCm39 genome assembly using BWA-MEM (v.0.7.17), filtered using samtools (v.1.9) and visualized using IGV (Integrative Genomics Viewer, Broad Institute, v.2.12.3).
Organoid proliferation assays
Single-cell suspensions of colon cells were generated as indicated above and embedded in 10 μl Matrigel domes at around 10 4 cells per dome in a 48-well plate. For each of the following 4 days, 220 μM resazurin (Sigma-Aldrich, R7017) was added to the culture medium and incubated for 4 h at 37 °C. Next, the resazurin-containing medium was collected and replaced with regular medium. Organoid proliferation was estimated by measuring the reduction of resazurin to fluorescent resorufin in the medium each day using the Tecan Infinite F500 microplate reader (Tecan) with 560 nm excitation and 590 nm emission filters. In the case of colony-formation assays, seeding was performed at around 10 3 cells per dome and the resulting colonies were counted after 3 days.
Organoid RNA extraction and bulk transcriptome profiling
Before RNA isolation, organoids were cultured for 3 days as indicated above and starved for 24 h in BM for the evaluation of growth-factor dependence. In the case of the Gpx2- knockdown experiments, 2 timepoints were analysed: 0 and 2 weeks after blue-light-induced activation (before and after oncogenic recombination, respectively). In all cases, cells were collected using TrypLE Express Enzyme as indicated above and lysed in RLT buffer (Qiagen, 74004), and the RNA was extracted using the QIAGEN RNeasy Micro Kit (Qiagen, 74004) according to the manufacturer’s instructions. Purified RNA was quality checked using a TapeStation 4200 (Agilent), and 500 ng was used for QuantSeq 3′ mRNA-seq library construction according to the manufacturer’s instructions (Lexogen, 015.96). Libraries were quality checked using a Fragment Analyzer (Agilent) and were sequenced in the NextSeq 500 (Illumina) system using NextSeq vm2.5 chemistry with Illumina protocol 15048776. Reads were aligned to the mouse genome (GRCm39) using star (v.2.7.0e) 43 . R (v.4.1.2) was used to perform the differential expression analyses. Count values were imported and processed using edgeR 44 . Expression values were normalized using the trimmed mean of M values (TMM) method 45 and low-expressed genes (<1 counts per million) were filtered out. Differentially expressed genes were identified using linear models (Limma-Voom) 46 and P values were adjusted for multiple comparisons using the Benjamini–Hochberg correction method 47 . Volcano plots and heat maps were generated using the EnhancedVolcano ( https://github.com/kevinblighe/EnhancedVolcano ) and heatmap3 ( https://github.com/slzhao/heatmap3 ) packages, respectively. The in vivo AKP signature was established from the differentially expressed genes between in vivo and organoid AKP lines with a log 2 -transformed fold change of at least |2|. To evaluate the enrichment of the in vivo AKP gene expression program across samples, the enrichment scores for both the upregulated and downregulated signatures were calculated using single-sample GSEA (ssGSEA) 48 . The difference between the two normalized enrichment scores yielded the fit score. ssGSEA was also used to analyse the enrichment of the MSigDB curated Hallmark gene set 49 in Gpx2 -knockdown organoids. Functional annotation was performed using DAVID 50 on the genes with a log 2 -transformed fold change of at least |1|. GOplot 51 was used for the integration of expression and functional annotation data. Known functional interactions among relevant genes were obtained through STRING 52 . Cytoscape 53 was used to perform network data integration and visualization.
Single-cell transcriptome profiling and lineage tracing
Lineage tracing was performed using the CellTag system 22 (V1 pooled barcode library, Addgene, 115643-LVC). In brief, we co-transduced inducible colon organoids with the CellTag barcode library (multiplicity of infection of around 5) and the OptoCre module as indicated above. These cells were then introduced and induced in the mini-colon system as indicated before. After 7 days in the system and when mini-colon tumours were clearly visible, we extracted the cells from mini-colons as indicated above. After pooling and filtering (40 μm) the cell suspensions from two mini-colons, the single-cell sequencing library was constructed using 10x Genomics Chromium 3′ reagents v3.1 according to the manufacturer’s instructions (10x Genomics, PN-1000269, PN-1000127, PN-1000215). Sequencing was performed using NovaSeq 6000 v1.5 reagents (Illumina protocol #1000000106351 v03) for around 100,000 reads per cell. The reads were aligned using Cell Ranger (v.6.1.2) 54 to the mouse genome (mm10) carrying artificial chromosomes for both GFP and CellTag UTR genes, as recommended by CellTag developers for facilitating barcode identification 55 . Raw count matrices were imported into R and analysed using Seurat (v.4.2.0) 56 . Dead cells were discarded on the basis of the number of detected genes (less than 3,000) and the percentage of mitochondrial genes (more than 20%), leading to 2,429 cells after filtering. The data were log-normalized and scaled, and dimensionality reduction was conducted using UMAP with 10 dimensions. Louvain clustering yielded 17 clusters that were merged and named on the basis of canonical cell type markers. Stem, cycling, progenitor, goblet and enteroendocrine cell scoring was based on published signatures in mini-intestines and in vivo 9 . Gene sets highlighting bottom, middle and top colonocytes were taken from enterocyte zonation studies 23 . Cancer stemness was scored based on the expression of Lgr5 , Cd44 and Sox9 . Intrinsic consensus molecular subtype (iCMS) signatures for colorectal cancer were obtained from published work 27 . Signature scoring was performed using burgertools ( https://github.com/nbroguiere/burgertools ). Visual representations of the data were generated using Seurat internal functions. For lineage-tracing analyses, CellTag detection, quantification and clone calling were performed as indicated by CellTag developers 55 , excluding cells expressing fewer than 2 or more than 30 CellTags. After filtering, 83 clonal populations were identified, from which only those with a minimum size of 5 cells were considered for further analyses. To identify clonal populations belonging to tumour cells, we looked for cells expressing transcripts carrying the genetically engineered Apc and Trp53 mutations, that is, deletions of exons 15 and 2–10, respectively (Extended Data Figs. 3g and 6b,c ). Note that this approach could not be performed for Kras , as the mutation is also present in the transcripts from WT cells (but not expressed). As scRNA-seq provides low coverage on exon junctions and therefore the presence of mutations can be assessed only in a small fraction of cells, we used both the cell-type composition and size distributions of bona fide mutationally confirmed tumour clonal populations to classify the rest of clones. Those falling within plus or minus 2 s.d. of the mean cell composition and size of bona fide tumours were classified as tumour clonal populations. Healthy clones were defined as those with a clearly distinct (outside the aforementioned range) cell type composition and the same upper limit size as was observed for tumour clones. After filtering and classification, 16 healthy and 18 tumour clonal populations were obtained and used for further analyses (Extended Data Fig. 6d ). To define the most robust tumour-clone-specific markers, the gene expression from cells in each clone was compared to that from cells in each other clone using the Wilcoxon rank-sum test. We considered only the positive markers and selected those with adjusted P < 10 −5 . The association of these markers with clinical parameters in patients with CRC (survival, lymph node staging) was performed through cBioPortal ( https://www.cbioportal.org/ ) using the 640-sample CRC TCGA dataset ( https://www.cancer.gov/tcga ) and a differential expression threshold equal or greater than |2 |. Further information is provided in the Data availability and Code availability sections.
shRNA-mediated transcript knockdown
Organoids were transduced as indicated above with lentiviral particles encoding Gpx2 shRNAs obtained from Sigma-Aldrich (TRCN0000076529, TRCN0000076531 and TRCN0000076532; sh Gpx2 1, sh Gpx2 2 and sh Gpx2 3, respectively) or, as a control, shRNA-free counterparts (Addgene, 65726). Transduced cells were selected with puromycin (5 μg ml −1 ; InvivoGen, ant-pr-1). Proper transcript knockdown was assessed using quantitative PCR with reverse transcription (RT–qPCR) and RNA-seq.
Analysis of mRNA abundance
Organoids were cultured and collected as indicated above. Cells were then lysed in RLT buffer and RNA was extracted using the QIAGEN RNeasy Micro Kit as indicated above. RT–qPCR was performed using the iTaq Universal SYBR Green One-Step Kit (Bio-Rad Laboratories, 1725150) and the QuantStudio 7 Flex Real-Time PCR System (Thermo Fisher Scientific, 4485701). Raw data were analysed using Design & Analysis Software (v.2.6.0, Thermo Fisher Scientific). We used the abundance of the endogenous Gapdh mRNA as internal normalization control. The following primers were used for transcript quantification: 5′-AGTTCGGACATCAGGAGAACTG-3′ (forward, Gpx2 ), 5′-GATGCTCGTTCTGCCCATTG-3′ (reverse, Gpx2 ), 5′-ATCCTGCACCACCAACTGCT-3′ (forward, Gapdh ) and 5′-GGGCCATCCACAGTCTTCTG-3′ (reverse, Gapdh ).
Microbiota and diet modelling
Inducible mini-colons were generated as indicated above. Once the epithelium was formed and before oncogenic induction, mini-colons were subjected to a conditioning period of 2 days in which luminal medium was (1) supplemented with 100 μM deoxycholate (Sigma-Aldrich, D2510), 10 mM butyrate (Sigma-Aldrich, B5887) or 10 mM β-hydroxybutyrate (Sigma-Aldrich, 54965); or (2) replaced with MEMα (calorie-restricted condition, Thermo Fisher Scientific, 22561-021) or Advanced DMEM/F12 supplemented with 30 μM palmitic acid (calorie-enriched condition, Sigma-Aldrich, P0500). The same concentrations were used in organoid control experiments, but these were added to the full culture medium as the luminal compartment is not accessible in organoids. To assess the relevance of luminal exposure to these factors in the mini-colon, the same total amounts were added in the basal medium reservoirs instead of the luminal channel. In all cases, after conditioning, oncogenic recombination was performed and the mini-colon was cultured as indicated above. The different medium compositions were replenished every day during luminal perfusion.
Statistics and reproducibility
The number of biological replicates ( n ), the type of statistical tests performed and the statistical significance for each experiment are indicated in the corresponding figure legend. For images associated with quantification charts (Fig. 1b,c with Fig. 1e ; Fig. 2b with Fig. 2c ; Fig. 2d with Extended Data Fig. 4b ; Fig. 2f with Fig. 2g ; Fig. 4b with Extended Data Fig. 9a ; Fig. 4i with Fig. 4j ; Extended Data Fig. 2a with Fig. 1e ; Extended Data Fig. 3d,e with Extended Data Fig. 3b ; Extended Data Fig. 3h with Fig. 1e ; Extended Data Fig. 5b with Extended Data Fig. 5c ; Extended Data Fig. 5d with Extended Data Fig. 5e ; Extended Data Fig. 9b with Extended Data Fig. 9c ; Extended Data Fig. 9e with Extended Data Fig. 9f ; Extended Data Fig. 9g with Fig. 4c ; Extended Data Fig. 9l with Extended Data Fig. 9m ; Extended Data Fig. 9n with Fig. 4g ; Extended Data Fig. 10c with Extended Data Fig. 10d ; Extended Data Fig. 10e with Extended Data Fig. 10f ), the number of replicates is the same as for the corresponding chart and is indicated in the figure legend of the latter. For the rest of representative images (Figs. 1d and 3h and Extended Data Figs. 1f , 2b,c,f , 3a , 4a , 7d , 9h and 10a,g–k ), three independent experiments were performed. scRNA-seq (Fig. 3a ) and exome sequencing with matched PCR (Extended Data Fig. 3f,g ) were performed with two independent sets of samples. Bulk RNA-seq was performed with at least three independent sets of samples. Unless otherwise indicated, statistical analyses were performed using GraphPad Prism v.9 (GraphPad). Data normality and equality of variances were analysed with Shapiro–Wilk and Bartlett’s tests, respectively. Parametric distributions were analysed using the Student’s t -test (when comparing two experimental groups) or ANOVA followed by either Dunnett’s test (when comparing more than two experimental groups with a single control group) or Tukey’s HSD test (when comparing more than two experimental groups with every other group). Nonparametric distributions were analysed using either Mann–Whitney U -tests (for comparisons of two experimental groups) or the Kruskal–Wallis followed by Dunn’s test (for comparisons of three or more than three experimental groups) tests. Sidak’s multiple-comparison test was used when comparing different sets of means. χ 2 tests were used to determine the significance of the differences between expected and observed frequencies. In all cases, values were considered to be significant when P ≤ 0.05. Data obtained are given as the mean ± s.e.m.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Bulk and single-cell RNA-seq data reported in this paper have been deposited at the Gene Expression Omnibus (GEO) public repository under accession number GSE221163 . The association analysis with clinical parameters in patients with CRC was performed through cBioPortal ( https://cbioportal.org ) using the 640-sample CRC TCGA dataset ( https://cancer.gov/tcga ). Source data are provided with this paper.
Code availability
The code used for data analysis is available at GitHub ( https://github.com/LorenzoLF/Mini-colon_bioengineering ) 57 and Zenodo ( https://doi.org/10.5281/zenodo.10057882 ) 58 .
Drost, J. & Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 18 , 407–418 (2018).
Article CAS PubMed Google Scholar
Rodrigues, J., Heinrich, M. A., Teixeira, L. M. & Prakash, J. 3D in vitro model (r)evolution: unveiling tumor-stroma interactions. Trends Cancer 7 , 249–264 (2021).
Tuveson, D. & Clevers, H. Cancer modeling meets human organoid technology. Science 364 , 952–955 (2019).
Article ADS CAS PubMed Google Scholar
Katt, M. E., Placone, A. L., Wong, A. D., Xu, Z. S. & Searson, P. C. In vitro tumor models: advantages, disadvantages, variables, and selecting the right platform. Front. Bioeng. Biotechnol. 4 , 12 (2016).
Article PubMed PubMed Central Google Scholar
Lee-Six, H. et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature 574 , 532–537 (2019).
Vendramin, R., Litchfield, K. & Swanton, C. Cancer evolution: Darwin and beyond. EMBO J. 40 , e108389 (2021).
Article CAS PubMed PubMed Central Google Scholar
Kim, J., Koo, B. K. & Knoblich, J. A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21 , 571–584 (2020).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459 , 262–265 (2009).
Nikolaev, M. et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 585 , 574–578 (2020).
Article ADS PubMed Google Scholar
Krotenberg Garcia, A. et al. Active elimination of intestinal cells drives oncogenic growth in organoids. Cell Rep 36 , 109307 (2021).
Liu, X. et al. Tumor-on-a-chip: from bioinspired design to biomedical application. Microsyst. Nanoeng. 7 , 50 (2021).
Article ADS PubMed PubMed Central Google Scholar
Augustine, R. et al. 3D bioprinted cancer models: revolutionizing personalized cancer therapy. Transl. Oncol. 14 , 101015 (2021).
Hubrecht, R. C. & Carter, E. The 3Rs and humane experimental technique: implementing change. Animals https://doi.org/10.3390/ani9100754 (2019).
Keum, N. & Giovannucci, E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 16 , 713–732 (2019).
Article PubMed Google Scholar
Bürtin, F., Mullins, C. S. & Linnebacher, M. Mouse models of colorectal cancer: past, present and future perspectives. World J. Gastroenterol. 26 , 1394–1426 (2020).
Drost, J. et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521 , 43–47 (2015).
Gehart, H. & Clevers, H. Tales from the crypt: new insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 16 , 19–34 (2019).
Levy, E. et al. Localization, function and regulation of the two intestinal fatty acid-binding protein types. Histochem. Cell Biol. 132 , 351–367 (2009).
Du, L. et al. CD44 is of functional importance for colorectal cancer stem cells. Clin. Cancer Res. 14 , 6751–6760 (2008).
Fleming, M., Ravula, S., Tatishchev, S. F. & Wang, H. L. Colorectal carcinoma: pathologic aspects. J. Gastrointest. Oncol. 3 , 153–173 (2012).
PubMed PubMed Central Google Scholar
Fujii, M. et al. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell 23 , 787–793 (2018).
Biddy, B. A. et al. Single-cell mapping of lineage and identity in direct reprogramming. Nature 564 , 219–224 (2018).
Article ADS CAS PubMed PubMed Central Google Scholar
Moor, A. E. et al. Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus axis. Cell 175 , 1156–1167 (2018).
Becker, W. R. et al. Single-cell analyses define a continuum of cell state and composition changes in the malignant transformation of polyps to colorectal cancer. Nat. Genet. 54 , 985–995 (2022).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143 , 134–144 (2010).
Kang, S. K. et al. Role of human aquaporin 5 in colorectal carcinogenesis. Am. J. Pathol. 173 , 518–525 (2008).
Joanito, I. et al. Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat. Genet. 54 , 963–975 (2022).
Hall, M. D. et al. Inhibition of glutathione peroxidase mediates the collateral sensitivity of multidrug-resistant cells to tiopronin. J. Biol. Chem. 289 , 21473–21489 (2014).
Louis, P., Hold, G. L. & Flint, H. J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 12 , 661–672 (2014).
Bernstein, C. et al. Carcinogenicity of deoxycholate, a secondary bile acid. Arch. Toxicol. 85 , 863–871 (2011).
Fu, T. et al. FXR regulates intestinal cancer stem cell proliferation. Cell 176 , 1098–1112 (2019).
Wu, X. et al. Effects of the intestinal microbial metabolite butyrate on the development of colorectal cancer. J. Cancer 9 , 2510–2517 (2018).
Dmitrieva-Posocco, O. et al. β-Hydroxybutyrate suppresses colorectal cancer. Nature 605 , 160–165 (2022).
Veettil, S. K. et al. Role of diet in colorectal cancer incidence: umbrella review of meta-analyses of prospective observational studies. JAMA Netw. Open 4 , e2037341 (2021).
Castejón, M. et al. Energy restriction and colorectal cancer: a call for additional research. Nutrients https://doi.org/10.3390/nu12010114 (2020).
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161 , 933–945 (2015).
Blutt, S. E. et al. Use of human tissue stem cell-derived organoid cultures to model enterohepatic circulation. Am. J. Physiol. Gastrointest. Liver Physiol. 321 , G270–G279 (2021).
Nikolaev, N. et al. Bioengineering microfluidic organoids-on-a-chip. Protocol Exchange https://doi.org/10.21203/rs.3.pex-903/v1 (2024).
Cho, K. R. & Vogelstein, B. Genetic alterations in the adenoma–carcinoma sequence. Cancer 70 , 1727–1731 (1992).
Meador, K. et al. Achieving tight control of a photoactivatable Cre recombinase gene switch: new design strategies and functional characterization in mammalian cells and rodent. Nucleic Acids Res. 47 , e97 (2019).
Wang, X., Chen, X. & Yang, Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods 9 , 266–269 (2012).
Sokolik, C. et al. Transcription factor competition allows embryonic stem cells to distinguish authentic signals from noise. Cell Syst. 1 , 117–129 (2015).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 , 15–21 (2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 , 139–140 (2010).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11 , R25 (2010).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15 , R29 (2014).
Reiner, A., Yekutieli, D. & Benjamini, Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19 , 368–375 (2003).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102 , 15545–15550 (2005).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) Hallmark gene set collection. Cell Syst. 1 , 417–425 (2015).
Dennis, G. et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4 , P3 (2003).
Walter, W., Sánchez-Cabo, F. & Ricote, M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 31 , 2912–2914 (2015).
Szklarczyk, D. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43 , D447–D452 (2015).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 , 2498–2504 (2003).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8 , 14049 (2017).
Kong, W. et al. CellTagging: combinatorial indexing to simultaneously map lineage and identity at single-cell resolution. Nat. Protoc. 15 , 750–772 (2020).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184 , 3573–3587 (2021).
Lorenzo-Martín, L. F. et al. Code for ‘Spatiotemporally resolved colorectal oncogenesis in mini-colons ex vivo’. GitHub github.com/LorenzoLF/Mini-colon_bioengineering (2024).
Lorenzo-Martín, L. F. et al. Code for ‘Spatiotemporally resolved colorectal oncogenesis in mini-colons ex vivo’. Zenodo https://doi.org/10.5281/zenodo.10057882 (2024).
Download references
Acknowledgements
We thank M. Thomson for the original light-inducible plasmids; M. Wernig and G. Neumayer for the initial idea and work on the doxycycline- and light-inducible system; C. Baur for discussing, designing and building the custom illumination device; A. Chrisnandy for assistance on photomask fabrication; O. Mitrofanova, B. Elci and Y. Tinguely for assistance on microdevice fabrication; D. Dutta and S. Li for input on organoid work; and J. Prébandier for administrative assistance. We acknowledge support from the following EPFL core facilities: CMi, CPG, PTBTG, HCF, BIOP, FCCF, BSF and GECF. This work was funded by the Swiss Cancer League (KFS-5103-08-2020), the Personalized Health and Related Technologies (PHRT) Initiative from the ETH Board and the EPFL.
Open access funding provided by EPFL Lausanne.
Author information
These authors contributed equally: L. Francisco Lorenzo-Martín, Tania Hübscher
Authors and Affiliations
Laboratory of Stem Cell Bioengineering, Institute of Bioengineering, School of Life Sciences and School of Engineering, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland
L. Francisco Lorenzo-Martín, Tania Hübscher, Nicolas Broguiere, Jakob Langer, Lucie Tillard & Matthias P. Lutolf
Swiss Institute for Experimental Cancer Research (ISREC), School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland
Amber D. Bowler & Freddy Radtke
Swiss Cancer Center Leman (SCCL), Lausanne, Switzerland
Institute of Human Biology (IHB), Roche Pharma Research and Early Development, Roche Innovation Center Basel, Basel, Switzerland
Mikhail Nikolaev & Matthias P. Lutolf
You can also search for this author in PubMed Google Scholar
Contributions
L.F.L.-M. conceived the study, designed experiments, performed the experimental and bioinformatic work, analysed the data, performed artwork design and wrote the manuscript. T.H. generated the OptoCre module and blue-light-associated systems, designed experiments, performed experimental work and analysed data. A.D.B. performed mouse-related work and isolated primary cells. N.B. performed bioinformatic work and analysed data. J.L. produced the microfluidic devices, optimized hydrogel patterning and generated mini-colon histological sections. L.T. performed experimental work. M.N. designed and developed the first mini-gut system. F.R. helped to conceive the work. M.P.L. conceived the work, designed experiments and edited the manuscript.
Corresponding authors
Correspondence to L. Francisco Lorenzo-Martín or Matthias P. Lutolf .
Ethics declarations
Competing interests.
The EPFL has filed for patent protection (EP16199677.2, PCT/EP2017/079651, US20190367872A1) on the scaffold-guided organoid technology used here, and M.P.L. and M.N. are named as inventors on those patents. M.P.L. is shareholder in Doppl, which is commercializing those patents. The other authors declare no competing interests.
Peer review
Peer review information.
Nature thanks Bradley Lega and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended data fig. 1 generation of blue–light-inducible akp colon organoids..
a , Schematic of the plasmids comprising the OptoCre module. The promoter, gene, and restriction sites used to generate the plasmids are indicated. b , Schematic of the integration of the different genetic elements that allow spatiotemporal control of oncogenic recombination in colon organoids. c , Schematic of the experimental workflow used to test and optimize the OptoCre system. d , Brightfield and fluorescence images of OptoCre-carrying inducible colon organoids exposed to the indicated conditions. Red and green signals correspond to healthy and mutated cells, respectively. Images were taken 48 h after induction. Scale bar, 200 μm. e , Recombination efficiency in inducible colon organoid exposed to the conditions indicated in panel d. *** P = 0.0001 (Kruskal-Wallis and Dunn’s multiple comparisons test; n = 9 cultures for each condition). Each point represents one well of organoids. Data represent mean ± SEM. f , Brightfield and fluorescence images of inducible colon organoids exposed to control (top) and activation (bottom) conditions, dissociated into single cells and replated in the absence of growth factors (BMGF medium). Green signal corresponds to mutated cells. Images were taken 24 h after replating. Scale bar, 200 μm.
Extended Data Fig. 2 Oncogenic mutations induce neoplastic growth in mini-colons.
a , Time-course brightfield and fluorescence images of non-induced healthy colon cells grown as conventional organoids and mini-colons. Absence of fluorescence signal indicates absence of oncogenic recombination. Scale bars, 200 μm (organoids) and 75 μm (mini-colons). b , Immunofluorescence images showing the expression of Fabp1 (left, green), and Sox9 (right, magenta) in healthy mini-colons cultured for 7 days. Scale bar, 100 μm. c , Fluorescence image (left) showing the presence of mutated cells 36 h after the blue–light-mediated induction of a mini-colon. The segmentation of each mutated cell is shown (right). Scale bar, 120 μm. d , Evolution of the number of mutated cells in an inducible mini-colon after blue–light-mediated activation. Each dot represents a measurement every 15 min. The 2 nd order smoothing of the data is shown. e , Time-course quantitation of cell shedding (total protein content) into the lumen of OptoCre and control mini-colons after blue-light induced oncogenic recombination. * P = 0.0156; ** P = 0.0033; *** P = 0.0003 (24 h), <0.0001 (96 h) (two-way ANOVA and Sidak’s multiple comparisons test; n = 3 mini-colons for each condition). Each point represents one mini-colon. Data represent mean ± SEM. f , Low- (left) and high-magnification (right) immunofluorescence images showing the presence of CD44 (magenta) and nuclei (blue) in a tumour-bearing mini-colon. White and grey arrowheads indicate early and advanced tumorigenic events, respectively. Scale bars, 120 μm (left) and 50 μm (right).
Extended Data Fig. 3 Oncogenic mutations induce full blown tumours in mini-colons.
a , Hematoxylin and eosin staining of a mini-colon tumour section. Scale bar, 25 μm. b , Time-course growth of tumours produced by cells derived from mini-colon tumours upon subcutaneous transplantation in immunodeficient mice ( n = 5 mice). As a reference, bona fide cancer cells from primary colon tumours are included. c , Image of the tumours at the endpoint of the experiment shown in panel b. d , Hematoxylin and eosin stainings of sections from the indicated tumour types. Zoomed-in areas (right) are indicated with a dashed square (left). Scale bar, 100 μm. e , Hematoxylin and eosin stainings of sections from mini-colon AKP implant tumours showing the presence of invading cancer cells (left, black arrowheads) and areas of cellular atypia (right, white arrowhead). Scale bar, 200 μm. f , Electrophoretic separation of PCR-amplified KRAS LSL locus in the indicated samples. See Methods for more details on PCR design. g , Whole exome sequencing coverage in the indicated loci and cells. Missing exons in recombined cells are indicated. h , Brightfield images of mini-colons of the indicated genotypes 23 days after blue light exposure. Neoplastic and tumour structures are indicated with black and white arrowheads in A and AK mini-colons, respectively. By that time tumours have extended throughout the whole mini-colon tissue in the case of the AKP model, forming a dense mass of cancer cells. Scale bar, 75 μm.
Extended Data Fig. 4 Mini-colon tumours display in vivo -like functional and transcriptional features.
a , Brightfield and fluorescence images of a mini-colon where blue light exposure has been targeted to a specific area (dashed blue line). Red and green signals correspond to healthy and mutated cells, respectively. Images were taken 36 h after induction. Scale bar, 75 μm. b , Multiplicity of tumours emerged in mini-colons cultured in the indicated conditions. * P = 0.0122; ** P = 0.0035; *** P = 0.0002 (two-way ANOVA and Sidak’s multiple comparisons test; n = 4 mini-colons for each condition). c , Distribution of tumour morphologies in mini-colons cultured in the indicated conditions. ** P = 0.0024 (two-way ANOVA and Sidak’s multiple comparisons test; n = 4 mini-colons for each condition). d , Brightfield images of the indicated colon organoid lines cultured for 3 days in full organoid medium. Scale bar, 200 μm. e , Metabolic activity (measured using resazurin) of the indicated colon organoid lines cultured in BMGF medium for the indicated time. Numeric labelling (1-8) is used to facilitate cell line identification. *** P = 0.0002 (mini-colon AKP #iii), <0.0001 (all other conditions) (two-way ANOVA and Sidak’s multiple comparisons test; n = 3 cultures for each line). f , Volcano plot of the differentially expressed genes between “in vivo” and “organoid AKP” cell lines. g , Top enriched functional clusters in the differentially expressed genes identified in panel f. h , Enrichment of the “in vivo AKP” transcriptional signature identified in panel f across the different “mini-colon” and “organoid AKP” lines. * P = 0.0137; ** P = 0.0032 (#iii), 0.0075 (#iv); *** P = 0.0001 (Brown-Forsythe ANOVA and Dunnett’s T3 multiple comparisons test (two-sided), n = 6 and 3 cultures for “organoid AKP” and the rest of cell lines, respectively). Each dot represents one culture. i , Main enriched functional terms in the differentially expressed genes between “mini-colon AKP” lines # i and # v. Significant terms are highlighted in red (one-sided Fisher’s exact test, adjusted P values). In b , c , e , and h , data represent mean ± SEM. GPL, glycerophospholipid; EL, ether lipid; PG, proteoglycans; SC, stem cell.
Extended Data Fig. 5 Tumorigenesis in the mini-colon leads to enhanced RTK signalling promoting growth factor independence.
a , Gene interaction network of the overlapping genes that are upregulated in AKP lines with high-proliferation potential in BM (“in vivo AKP”, “mini-colon AKP” #v) when compared to low-growth counterparts (“organoid AKP”, “mini-colon AKP” #i). Network hubs are highlighted with circles. b , Brightfield images of “mini-colon AKP” #v organoids cultured for 3 days with the indicated media and compounds. Scale bar, 200 μm. c , Metabolic activity (measured using resazurin) of “mini-colon AKP” #v organoids cultured in the indicated conditions. * P = 0.0236; *** P < 0.0001 (two-way ANOVA and Sidak’s multiple comparisons test; n = 4 cultures for infigranib and 8 for the rest of conditions). d , Brightfield images of “organoid AKP” cells cultured for 3 days in BM with the indicated growth factors. Scale bar, 200 μm. e , Metabolic activity (measured using resazurin) of “organoid AKP” cells cultured in the indicated conditions. *** P < 0.0001 (two-way ANOVA and Sidak’s multiple comparisons test; n = 8 cultures for each condition). In c and e , each point represents one well of organoids and data represent mean ± SEM.
Extended Data Fig. 6 Mini-colons comprise a complex cellular ecosystem.
a , Expression distribution of cell–type-specific markers across mini-colon cells. Cell-type labels can be found in Fig. 3b . b , Examples of single-cell RNA reads capturing exon-exon junctions that reveal the expected oncogenic recombination in Apc . c , Examples of single-cell RNA reads capturing exon-exon junctions that reveal the expected oncogenic recombination in Trp53 . d , Unsupervised UMAP clustering of the main cell types found in each of the healthy and tumour clonal populations found within the mini-colon. Tumour clones carry the “CRC” label. UMAP structure corresponds to the one shown in Fig. 3b .
Extended Data Fig. 7 Mini-colons display intra- and inter-tumour diversity.
a , Expression distribution of proliferation ( Mki67 ), stemness ( Cd44 ), and differentiation ( Krt20 ) markers within a single clonal tumour population. b , Heatmap of the genes showing the strongest ( P < 10 −5 ) differential expression across mini-colon tumours. The tumour clonal population is indicated on top. c , Expression of the indicated genes in the indicated tumour clones. *** P = 2.74·10 −14 ( Slpi , clone #1), 4.06·10 −50 ( Prdm16 , clone #14), 1.05·10 −13 ( Aqp5 , clone #25) (two-sided Wilcoxon rank-sum test; n = 540 cells). Each point represents one cell. d , Immunofluorescence images showing the expression of Il1a (left, green), Cdkn2a (right, red), and the presence of nuclei (cyan) in tumour-bearing and control mini-colons. White and grey arrowheads indicate positive and negative tumours, respectively, in terms of marker expression. Scale bar, 100 μm.
Extended Data Fig. 8 The tumour heterogeneity in the mini-colon is relatable to the human context.
a , Expression distribution of intrinsic consensus molecular subtype (iCMS) signatures across tumour cells in the mini-colon. Cell-type labels can be found in Fig. 3d . Exp, expression. b , Fraction of cells within each tumour clone in the mini-colon classified in each iCMS group. c , Survival of CRC patients from the TCGA database according to the expression of the indicated tumour clone-specific markers. The logrank test P value is indicated ( n = 375 patients). d , Presence of cancer cells in lymph nodes from CRC patients from the TCGA database according to the expression of the indicated tumour clone-specific markers. *** P = 2.714·10 −5 (two-sided Wilcoxon test; n = 375 patients). e , Lymph node staging in CRC patients from the TCGA database according to the expression of the indicated tumour clone-specific markers. * P = 0.0222; ** P = 4.923·10 −3 (two-sided Chi-squared test; n = 375 patients).
Extended Data Fig. 9 Gpx2 regulates colonocyte stemness and tumorigenesis.
a , Multiplicity of tumours emerged in mini-colons treated with the indicated compound upon oncogenic induction. *** P = 0.0001 (day 5), <0.0001 (all other conditions) (two-way ANOVA and Sidak’s multiple comparisons test; n = 6 mini-colons for each condition). b , Brightfield images of colon organoids treated with the indicated compound after tumorigenic recombination. Images correspond to 3 days after induction. Scale bar, 200 μm. c , Metabolic activity (measured using resazurin) of organoids cultured in the indicated conditions and times after oncogenic recombination. No significant differences (two-way ANOVA and Sidak’s multiple comparisons test; n = 4 cultures for each condition). d , qRT-PCR based quantitation of Gpx2 mRNA in the indicated cell lines. *** P < 0.0001 (one-way ANOVA and Tukey’s multiple comparisons test; n = 6, 4, and 3 organoid cultures for parental, sh Gpx2 #1, and the rest of the lines, respectively). e , Brightfield images of non-induced colon organoids of the indicated genotypes after 3 days of culture. Scale bar, 200 μm. f , Metabolic activity (measured using resazurin) of non-induced colon organoids of the indicated genotypes at the indicated times. No significant differences (two-way ANOVA and Sidak’s multiple comparisons test; n = 6 cultures for each condition). g , Brightfield images of mini-colons of the indicated genotypes after tumorigenic recombination. Images correspond to 6 days after induction. Scale bar, 75 μm. h , Brightfield images of colon organoids of the indicated genotypes after tumorigenic recombination. Images correspond to 6 days after induction. Scale bar, 200 μm. i , Volcano plot showing the differentially expressed genes upon Gpx2 knockdown in non-transformed colon cells. j , Bubble plot showing the main enriched functional terms in the differentially expressed genes upon Gpx2 knockdown in non-transformed colon cells. Significant terms are highlighted in either blue (downmodulated) or red (upmodulated) (one-sided Fisher’s exact test, adjusted P values). k , Enrichment of the indicated hallmark signatures from the MSigDB in the indicated cell lines. ** P = 0.0013; *** P = 0.0008 (Wnt), 0.0003 (EMT, before recombination), <0.0001 (all other conditions); NS, not significant (one-way ANOVA and Tukey’s multiple comparisons test; n = 3 cultures for each condition). l , Colony assay images of non-induced colon organoids of the indicated genotypes after 3 days of culture in the indicated media conditions. Scale bar, 200 μm. m , Clonogenic capacity of non-induced colon organoids of the indicated genotypes after 3 days of culture in the indicated media conditions. * P = 0.0219; ** P = 0.0012 (EN, sh Gpx2 #3), 0.0036 (BMGF, sh Gpx2 #1); *** P < 0.0001 (two-way ANOVA and Dunnet’s multiple comparisons test; n = 3 cultures for each condition). n , Brightfield images of Gpx2 knockdown mini-colons that had undergone the indicated pre-treatment before tumorigenic recombination. Images correspond to 7 days after tumour induction. Scale bar, 75 μm. In a , c , d , f , k , and m , each point represents one well of organoids and data represent mean ± SEM.
Extended Data Fig. 10 Mini-colons provide experimental versatility and resolution to tumorigenic studies.
a , Brightfield images of colon organoids treated with the indicated bacterial metabolites. Images correspond to 5 days after oncogenic induction. Scale bar, 200 μm. b , Schematic of the experimental setup used to evaluate the relevance of luminal access in tumorigenic studies. c , Brightfield images of mini-colons treated with the indicated diets according to experimental setup displayed in panel b (left). Images correspond to 6 days after tumorigenic induction. Scale bar, 75 μm. d , Multiplicity of tumours emerged in mini-colons treated with the indicated diets according to experimental setup displayed in panel b (left). *** P = 0.0001 (day 7), <0.0001 (days 6 and 8) (two-way ANOVA and Sidak’s multiple comparisons test; n = 4 and 3 mini-colons for calorie-restricted and -enriched diets, respectively). e , Brightfield images of mini-colons treated with the indicated diets according to experimental setup displayed in panel b (right). Images correspond to 6 days after tumorigenic induction. Scale bar, 75 μm. f , Multiplicity of tumours emerged in mini-colons treated with the indicated diets according to experimental setup displayed in panel b (right). Differences are not significant (two-way ANOVA and Sidak’s multiple comparisons test; n = 4 and 3 mini-colons for calorie-restricted and -enriched diets, respectively). g , Brightfield image of a healthy (non-transformed) mini-colon with integrated stromal cells in the extracellular matrix. Scale bar, 75 μm. h , Immunofluorescence image showing the presence of E-cadherin (green) and Vimentin (magenta) in the mini-colon shown in panel g. Scale bar, 75 μm. i , Brightfield images of mini-colons in the indicated mono- (left) and co-culture (right) setups. Images correspond to 6 days after tumorigenic induction. Scale bar, 75 μm. j , Brightfield image of an invasive front (arrowhead) formed in response to the presence of stromal cells in a mini-colon. The image corresponds to 6 days after tumorigenic induction (zoomed-in from panel i). Scale bar, 30 μm. k , Immunofluorescence image showing the presence of E-cadherin (green) and Vimentin (magenta) in the invasive front from panel j 23 days after tumorigenic induction. Scale bar, 20 μm. In d and f , data represent mean ± SEM.
Supplementary information
Reporting summary, supplementary table 1.
Differentially expressed genes in shGpx2 colon organoids before oncogenic recombination.
Supplementary Table 2
Differentially expressed genes in shGpx2 colon organoids after oncogenic recombination
Supplementary Video 1
Early response to oncogenic activation within a mini-colon. 46 h time-lapse video of mutated cells in a mini-colon 24 h after oncogenic recombination.
Supplementary Video 2
Hyperplasia and early tumour development in a mini-colon. 36 h time-lapse video of a mini-colon with multiple tumour-initiating events 5 days after oncogenic recombination.
Supplementary Video 3
Ex vivo tumour development in a mini-colon. 38 h time-lapse video of tumour development in a mini-colon 9 days after oncogenic recombination.
Supplementary Video 4
Cancer stem cells initiate tumour development in mini-colons. 3D visualization of cancer stem cell marker CD44 overexpression in early tumorigenic sites.
Supplementary Video 5
Intratumour complexity in mini-colons. 3D visualization of CD44 (cancer stem cell marker) and FABP1 (mature colonocyte marker) expression in mini-colon tumours and epithelium.
Source data
Source data fig. 1, source data fig. 2, source data fig. 3, source data fig. 4, source data extended data fig. 1, source data extended data fig. 2, source data extended data fig. 3, source data extended data fig. 4, source data extended data fig. 5, source data extended data fig. 8, source data extended data fig. 9, source data extended data fig. 10, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Lorenzo-Martín, L.F., Hübscher, T., Bowler, A.D. et al. Spatiotemporally resolved colorectal oncogenesis in mini-colons ex vivo. Nature (2024). https://doi.org/10.1038/s41586-024-07330-2
Download citation
Received : 02 February 2023
Accepted : 18 March 2024
Published : 24 April 2024
DOI : https://doi.org/10.1038/s41586-024-07330-2
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Mini-colon and brain 'organoids' shed light on cancer and other diseases.
- Sara Reardon
Nature (2024)
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Cancer newsletter — what matters in cancer research, free to your inbox weekly.
- help_outline help
iRubric: The Mini-Research Paper rubric
- Research paper - emphasis on relevance of research
, , .
.
.

AI + Machine Learning , Announcements , Azure AI , Azure AI Studio
Introducing Phi-3: Redefining what’s possible with SLMs
By Misha Bilenko Corporate Vice President, Microsoft GenAI
Posted on April 23, 2024 4 min read
- Tag: Copilot
- Tag: Generative AI
We are excited to introduce Phi-3, a family of open AI models developed by Microsoft. Phi-3 models are the most capable and cost-effective small language models (SLMs) available, outperforming models of the same size and next size up across a variety of language, reasoning, coding, and math benchmarks. This release expands the selection of high-quality models for customers, offering more practical choices as they compose and build generative AI applications.
Starting today, Phi-3-mini , a 3.8B language model is available on Microsoft Azure AI Studio , Hugging Face , and Ollama .
- Phi-3-mini is available in two context-length variants—4K and 128K tokens. It is the first model in its class to support a context window of up to 128K tokens, with little impact on quality.
- It is instruction-tuned, meaning that it’s trained to follow different types of instructions reflecting how people normally communicate. This ensures the model is ready to use out-of-the-box.
- It is available on Azure AI to take advantage of the deploy-eval-finetune toolchain, and is available on Ollama for developers to run locally on their laptops.
- It has been optimized for ONNX Runtime with support for Windows DirectML along with cross-platform support across graphics processing unit (GPU), CPU, and even mobile hardware.
- It is also available as an NVIDIA NIM microservice with a standard API interface that can be deployed anywhere. And has been optimized for NVIDIA GPUs .
In the coming weeks, additional models will be added to Phi-3 family to offer customers even more flexibility across the quality-cost curve. Phi-3-small (7B) and Phi-3-medium (14B) will be available in the Azure AI model catalog and other model gardens shortly.
Microsoft continues to offer the best models across the quality-cost curve and today’s Phi-3 release expands the selection of models with state-of-the-art small models.

Azure AI Studio
Phi-3-mini is now available
Groundbreaking performance at a small size
Phi-3 models significantly outperform language models of the same and larger sizes on key benchmarks (see benchmark numbers below, higher is better). Phi-3-mini does better than models twice its size, and Phi-3-small and Phi-3-medium outperform much larger models, including GPT-3.5T.
All reported numbers are produced with the same pipeline to ensure that the numbers are comparable. As a result, these numbers may differ from other published numbers due to slight differences in the evaluation methodology. More details on benchmarks are provided in our technical paper .
Note: Phi-3 models do not perform as well on factual knowledge benchmarks (such as TriviaQA) as the smaller model size results in less capacity to retain facts.
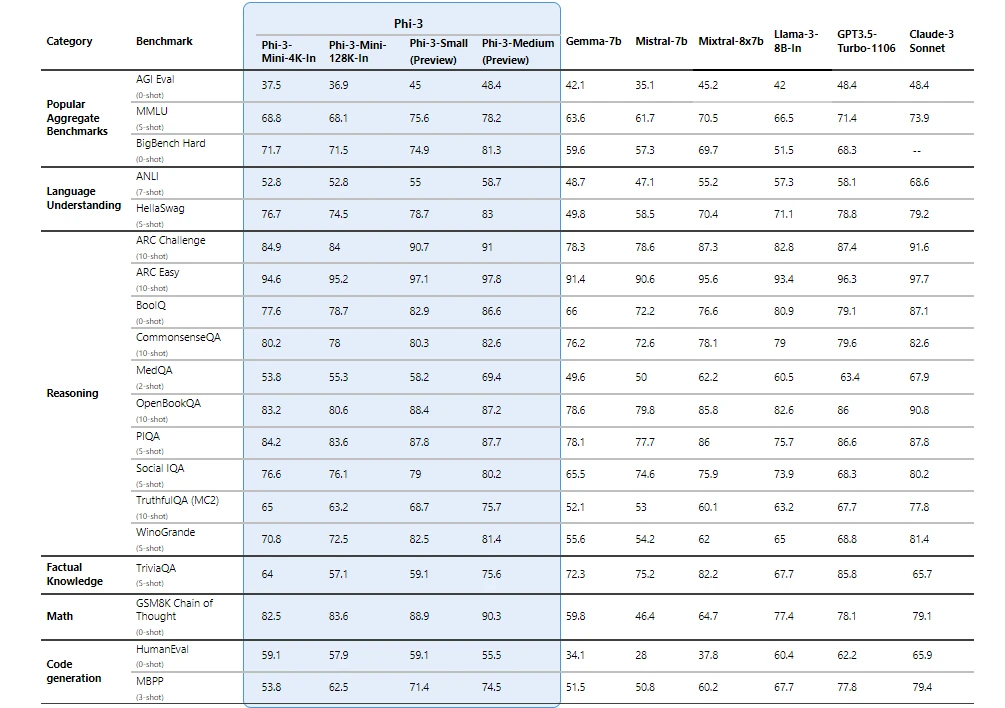
Safety-first model design
Responsible ai principles
Phi-3 models were developed in accordance with the Microsoft Responsible AI Standard , which is a company-wide set of requirements based on the following six principles: accountability, transparency, fairness, reliability and safety, privacy and security, and inclusiveness. Phi-3 models underwent rigorous safety measurement and evaluation, red-teaming, sensitive use review, and adherence to security guidance to help ensure that these models are responsibly developed, tested, and deployed in alignment with Microsoft’s standards and best practices.
Building on our prior work with Phi models (“ Textbooks Are All You Need ”), Phi-3 models are also trained using high-quality data. They were further improved with extensive safety post-training, including reinforcement learning from human feedback (RLHF), automated testing and evaluations across dozens of harm categories, and manual red-teaming. Our approach to safety training and evaluations are detailed in our technical paper , and we outline recommended uses and limitations in the model cards. See the model card collection .
Unlocking new capabilities
Microsoft’s experience shipping copilots and enabling customers to transform their businesses with generative AI using Azure AI has highlighted the growing need for different-size models across the quality-cost curve for different tasks. Small language models, like Phi-3, are especially great for:
- Resource constrained environments including on-device and offline inference scenarios.
- Latency bound scenarios where fast response times are critical.
- Cost constrained use cases, particularly those with simpler tasks.
For more on small language models, see our Microsoft Source Blog .
Thanks to their smaller size, Phi-3 models can be used in compute-limited inference environments. Phi-3-mini, in particular, can be used on-device, especially when further optimized with ONNX Runtime for cross-platform availability. The smaller size of Phi-3 models also makes fine-tuning or customization easier and more affordable. In addition, their lower computational needs make them a lower cost option with much better latency. The longer context window enables taking in and reasoning over large text content—documents, web pages, code, and more. Phi-3-mini demonstrates strong reasoning and logic capabilities, making it a good candidate for analytical tasks.
Customers are already building solutions with Phi-3. One example where Phi-3 is already demonstrating value is in agriculture, where internet might not be readily accessible. Powerful small models like Phi-3 along with Microsoft copilot templates are available to farmers at the point of need and provide the additional benefit of running at reduced cost, making AI technologies even more accessible.
ITC, a leading business conglomerate based in India, is leveraging Phi-3 as part of their continued collaboration with Microsoft on the copilot for Krishi Mitra, a farmer-facing app that reaches over a million farmers.
“ Our goal with the Krishi Mitra copilot is to improve efficiency while maintaining the accuracy of a large language model. We are excited to partner with Microsoft on using fine-tuned versions of Phi-3 to meet both our goals—efficiency and accuracy! ” Saif Naik, Head of Technology, ITCMAARS
Originating in Microsoft Research, Phi models have been broadly used, with Phi-2 downloaded over 2 million times. The Phi series of models have achieved remarkable performance with strategic data curation and innovative scaling. Starting with Phi-1, a model used for Python coding, to Phi-1.5, enhancing reasoning and understanding, and then to Phi-2, a 2.7 billion-parameter model outperforming those up to 25 times its size in language comprehension. 1 Each iteration has leveraged high-quality training data and knowledge transfer techniques to challenge conventional scaling laws.
Get started today
To experience Phi-3 for yourself, start with playing with the model on Azure AI Playground . You can also find the model on the Hugging Chat playground . Start building with and customizing Phi-3 for your scenarios using the Azure AI Studio . Join us to learn more about Phi-3 during a special live stream of the AI Show.
1 Microsoft Research Blog, Phi-2: The surprising power of small language models, December 12, 2023 .
Let us know what you think of Azure and what you would like to see in the future.
Provide feedback
Build your cloud computing and Azure skills with free courses by Microsoft Learn.
Explore Azure learning
Related posts
AI + Machine Learning , Azure AI , Azure AI Content Safety , Azure Cognitive Search , Azure Kubernetes Service (AKS) , Azure OpenAI Service , Customer stories
AI-powered dialogues: Global telecommunications with Azure OpenAI Service chevron_right
AI + Machine Learning , Azure AI , Azure AI Content Safety , Azure OpenAI Service , Customer stories
Generative AI and the path to personalized medicine with Microsoft Azure chevron_right
AI + Machine Learning , Azure AI , Azure AI Services , Azure AI Studio , Azure OpenAI Service , Best practices
AI study guide: The no-cost tools from Microsoft to jump start your generative AI journey chevron_right
AI + Machine Learning , Azure AI , Azure VMware Solution , Events , Microsoft Copilot for Azure , Microsoft Defender for Cloud
Get ready for AI at the Migrate to Innovate digital event chevron_right

IMAGES
VIDEO
COMMENTS
Do Review Papers Have Abstracts? • Yes, but there are two kinds of abstract: - Informative: Used for research reports. They summarize the study, including the findings and conclusions. - Descriptive: Used for reviews. They summarize the subject of the review and the approach the reviewer has taken in his or her coverage of the subject.
In this work, we introduce Mini-Gemini, a simple and effective framework enhancing multi-modality Vision Language Models (VLMs). Despite the advancements in VLMs facilitating basic visual dialog and reasoning, a performance gap persists compared to advanced models like GPT-4 and Gemini. We try to narrow the gap by mining the potential of VLMs for better performance and any-to-any workflow from ...
Mini Reviews are peer-reviewed, have a maximum word count of 3,000 and may contain no more than 2 Figures/Tables. Authors are required to pay a fee (B-type article) to publish a Mini Review. The collection of review articles from this Research Topic will serve as literature references to a new course on the Principles of Neuroscience.
Published 16 October, 2023. A mini research paper outline is a great way to organize your thoughts and get started on an assignment. This blog post is going to walk you through the process of writing a mini-research paper outline. It will not only help you with your own work but also give insight into what professors are looking for from their ...
In this work, we introduce Mini-Gemini, a simple and effective framework enhancing multi-modality Vision Language Models (VLMs). Despite the advancements in VLMs facilitating basic visual dialog and reasoning, a performance gap persists compared to advanced models like GPT-4 and Gemini. We try to narrow the gap by mining the potential of VLMs ...
JELL (Journal of English Language and literature) STIBA IEC Jakarta Volume 8, Issue 2, September 2023 261 p-ISSN 2540-8216, e-ISSN 2654-3745
The paper uses Derrida's deconstruction theory to read and engage with the discourse on curriculum integration in these documents: Curriculum and Assessment Policy (2009), Guide to Continuous ...
The IEEE Internet of Things Magazine (IEEE IoTM) is soliciting articles for its mini-series on "AI for IoT". The proliferation of IoT devices and sensors, coupled with advancements in AI algorithms and computing technologies, has paved the way for a new era of intelligent IoT systems. AI techniques are increasingly integrated into IoT architectures to enable advanced analytics, autonomous ...
If you find this repo useful for your research, please consider citing the paper @article{li2024mgm, title={Mini-Gemini: Mining the Potential of Multi-modality Vision Language Models}, author={Li, Yanwei and Zhang, Yuechen and Wang, Chengyao and Zhong, Zhisheng and Chen, Yixin and Chu, Ruihang and Liu, Shaoteng and Jia, Jiaya}, journal={arXiv ...
SUBMIT PAPER. Journalism & Mass Communication Quarterly. Impact Factor: 3.6 / 5-Year Impact Factor: 4.5 . ... resistance, and representing national values in the Brazilian television mini-series. Show details Hide details. Niall P Brennan. Media, Culture & Society. Mar 2015. ... Sage Research Methods Supercharging research opens in new tab;
width of 600 pixels (standard), 1200 pixels (high resolution). Figures in the final PDF version: width of 85 mm for half page width figure. width of 170 mm for full page width figure. maximum height of 225 mm for figure and legend. image resolution of approximately 300 dpi (dots per inch) at the final size.
What is a mini-review. A mini-review (like a book review) is simply the careful summary of a particular scholar's work. It is usually a 2-3 page articulation of a work's main thesis, supporting arguments and major points. After this is complete, a final step in the mini-review is to take the content of 1-2 pages and shrink that down to one ...
Ideally, a literature review should not identify as a major research gap an issue that has just been addressed in a series of papers in press (the same applies, of course, to older, overlooked studies ("sleeping beauties" )). This implies that literature reviewers would do well to keep an eye on electronic lists of papers in press, given ...
Choose a research paper topic. Conduct preliminary research. Develop a thesis statement. Create a research paper outline. Write a first draft of the research paper. Write the introduction. Write a compelling body of text. Write the conclusion. The second draft.
The timeseries data are transformed into spatial data in mini-batches that consist of multivariate sensor data in each box. Flow rates (ton/h) of Crude Oil and three main branches of by-products ...
A series of SLMs offer many of the same capabilities found in LLMs but are smaller in size and are trained on smaller amounts of data. ... That led the Microsoft Research machine learning expert to wonder how much an AI model could learn using only words a 4-year-old could understand - and ultimately to an innovative training approach that ...
It undoubtedly helps the mini-series that the whole group — the original members Mr. Brown, Michael Bivins, Mr. Tresvant, Ricky Bell and Ronnie DeVoe, as well as Mr. Gill, who joined after Mr ...
The Journal of Politics, 67 (4), 1099-1121. This research paper, "Drug Addiction Issues in The Corner Miniseries" is published exclusively on IvyPanda's free essay examples database. You can use it for research and reference purposes to write your own paper. However, you must cite it accordingly . Donate a paper.
This special i,sue nn bullying and victimization in Schooll',I'rc!l%gr Rel'ieu' highlights current research elTol1:> in American schools on bullying and peer victimization. and how this research can inform prevention and intervention planning. nlis introductory article provides a brief overview of several major insights gained over the last decade from research on bullying in school-aged youth ...
We introduce phi-3-mini, a 3.8 billion parameter language model trained on 3.3 trillion tokens, whose overall performance, as measured by both academic benchmarks and internal testing, rivals that of models such as Mixtral 8x7B and GPT-3.5 (e.g., phi-3-mini achieves 69% on MMLU and 8.38 on MT-bench), despite being small enough to be deployed on a phone. The innovation lies entirely in our ...
iRubric M293X4: The mini-research paper should be approximately 3 pages in length, 1.5 spaces, and in 12 pt. Times New Roman font. It should also follow MLA format/citation as discussed in class. You must submit your annotated source notes with your paper.. Free rubric builder and assessment tools.
Abstract. Several models have been proposed to connect academia and practice in order to improve long-term care. In this paper we propose and describe the "Mini-Research Group" as an alternative model of such collaboration. The formation of mini-research groups was the unplanned by-product of a longitudinal action research project headed by ...
Three-dimensional organoid culture technologies have revolutionized cancer research by allowing for more realistic and scalable reproductions of both tumour and microenvironmental structures1-3.
iRubric J2282A6: Research the approved topic. Submit one 4-5 page typed paper, double spaced, 12 point font, Times New Roman or Arial style font, following MLA guidelines. Include a Works Cited page citing at least 3 online sources used to research the paper. (Only one direct quote allowed and cannot be more than three lines) Online sources must include articles from websites, online articles ...
Build the future of AI with Meta Llama 3. Now available with both 8B and 70B pretrained and instruction-tuned versions to support a wide range of applications.
Thanks to their smaller size, Phi-3 models can be used in compute-limited inference environments. Phi-3-mini, in particular, can be used on-device, especially when further optimized with ONNX Runtime for cross-platform availability. The smaller size of Phi-3 models also makes fine-tuning or customization easier and more affordable.
Sample research for students. chapter the problem and its setting introduction in our society today, depression is becoming pressing issue on childcare. Skip to document. ... In this mini-research, the researcher chose to include all Grade 10 students of a certain school with a total population of 40 students.