web development Recently Published Documents
Total documents.
- Latest Documents
- Most Cited Documents
- Contributed Authors
- Related Sources
- Related Keywords

Website Developmemt Technologies: A Review
Abstract: Service Science is that the basis of knowledge system and net services that judge to the provider/client model. This paper developments a technique which will be utilized in the event of net services like websites, net applications and eCommerce. The goal is to development a technique that may add structure to a extremely unstructured drawback to help within the development and success of net services. The new methodology projected are going to be referred to as {the net|the online|the net} Development Life Cycle (WDLC) and tailored from existing methodologies and applied to the context of web development. This paper can define well the projected phases of the WDLC. Keywords: Web Development, Application Development, Technologies, eCommerce.
Analysis of Russian Segment of the Web Development Market Operating Online on Upwork
The Russian segment of the web services market in the online environment, on the platform of the Upwork freelance exchange, is considered, its key characteristics, the composition of participants, development trends are highlighted, and the market structure is identified. It is found that despite the low barriers to entry, the web development market is very stable, since the composition of entrenched firms that have been operating for more than six years remains. The pricing policy of most Russian companies indicates that they work in the middle price segment and have low budgets, which is due to the specifics of the foreign market and high competition.
Farming Assistant Web Services: Agricultor
Abstract: Our farming assistant web services provides assistance to new as well as establish farmers to get the solutions to dayto-day problems faced in the field. A farmer gets to connect with other farmers throughout India to get more information about a particular crop which is popular in other states. Keywords: Farmers, Assistance, Web Development
Tradução de ementas e histórico escolar para o inglês: contribuição para participação de discentes do curso técnico em informática para internet integrado ao ensino médio em programas de mobilidade acadêmica / Translation of summary and school records into english: contribution to the participation of high school with associate technical degree on web development students in academic mobility programs
Coded websites vs wordpress websites.
This document gives multiple instructions related to web developers using older as well as newer technology. Websites are being created using newer technologies like wordpress whereas on the other hand many people prefer making websites using the traditional way. This document will clear the doubt whether an individual should use wordpress websites or coded websites according to the users convenience. The Responsiveness of the websites, the use of CMS nowadays, more and more up gradation of technologies with SEO, themes, templates, etc. make things like web development much much easier. The aesthetics, the culture, the expressions, the features all together add up in order make the designing and development a lot more efficient and effective. Digital Marketing has a tremendous growth over the last two years and yet shows no signs of stopping, is closely related with the web development environment. Nowadays all businesses are going online due to which the impact of web development has become such that it has become an integral part of any online business.
Cognitive disabilities and web accessibility: a survey into the Brazilian web development community
Cognitive disabilities include a diversity of conditions related to cognitive functions, such as reading, understanding, learning, solving problems, memorization and speaking. They differ largely from each other, making them a heterogeneous complex set of disabilities. Although the awareness about cognitive disabilities has been increasing in the last few years, it is still less than necessary compared to other disabilities. The need for an investigation about this issue is part of the agenda of the Challenge 2 (Accessibility and Digital Inclusion) from GranDIHC-Br. This paper describes the results of an online exploratory survey conducted with 105 web development professionals from different sectors to understand their knowledge and barriers regarding accessibility for people with cognitive disabilities. The results evidenced three biases that potentially prevent those professionals from approaching cogni-tive disabilities: strong organizational barriers; difficulty to understand user needs related to cognitive disabilities; a knowledge gap about web accessibility principles and guidelines. Our results confirmed that web development professionals are unaware about cognitive disabilities mostly by a lack of knowledge about them, even if they understand web accessibility in a technical level. Therefore, we suggest that applied research studies focus on how to fill this knowledge gap before providing tools, artifacts or frameworks.
PERANCANGAN WEB RESPONSIVE UNTUK SISTEM INFORMASI OBAT-OBATAN
A good information system must not only be neat, effective, and resilient, but also must be user friendly and up to date. In a sense, it is able to be applied to various types of electronic devices, easily accessible at any whereand time (real time), and can be modified according to user needs in a relatively easy and simple way. Information systems are now needed by various parties, especially in the field of administration and sale of medicines for Cut Nyak Dhien Hospital. During this time, recording in books has been very ineffective and caused many problems, such as difficulty in accessing old data, asa well as the information obtained was not real time. To solve it, this research raises the theme of the appropriate information system design for the hospital concerned, by utilizing CSS Bootstrap framework and research methodology for web development, namely Web Development Life Cycle. This research resulted in a responsive system by providing easy access through desktop computers, tablets, and smartphones so that it would help the hospital in the data processing process in real time.
Web Development and performance comparison of Web Development Technologies in Node.js and Python
“tom had us all doing front-end web development”: a nostalgic (re)imagining of myspace, assessment of site classifications according to layout type in web development, export citation format, share document.
Initial framework for website design and development
- Original Research
- Published: 23 October 2017
- Volume 9 , pages 363–375, ( 2017 )
Cite this article

- Jatinder Manhas 1
2245 Accesses
8 Citations
Explore all metrics
Website development is becoming a growing concern for all organizations in this world. However the process and lifecycle involved in their development is still uncertain. The Web based system development usually involves more heterogeneous stakeholders as compared to traditional software’s development construction. The process models used for the development of websites stands borrowed from the traditional software development approaches. The process of website development involves several fields of web engineering and multimedia applications which require altogether a different skill set and development processes. The growth of web based system has been exponential. An estimated size of public web and deep web has reached to 40 billion pages and where the pages are assembled on the fly in response to the user requests touches between 400 and 750 billion pages, respectively. The web based system size is continuously expanding like the universe after the big bang. Many researchers and practitioners have noted the rapid growth of internet for commercial purposes since its birth in 1990’s. The web presence allows small companies to compete comparatively at par with large enterprises which increases keenness for every organization to develop a website. An increase in the amount of web development work, forces the designers/webmasters working in different website development houses to carry out work in a well planned and systematic manner. The concerned organizations and different web development houses must, however follow specific methodologies/standards/guidelines recommended by various organizations for advancement in present web development process models. The adoption of traditional methodologies of web development leads to the production of poorly designed web applications which show variable performance across different browsing platforms and have high probability failure rate. This propagates in the lack of confidence during web development and ultimately moves towards Web-Crisis. In order to avoid Web-Crises, there is a desperate need of designing and developing the newer process models that are especially concerned with website development. As such there is a need of disciplined approach towards the development of web-based systems. Researches also suggest that there is a non uniform approach in web based systems development. The authors suggest that a specialized model for web development needs to be designed and developed to satisfy the dynamic categories of the stakeholders present worldwide. This paper discussed about the various traditional software and web development models popularly used in developing web applications and their suitability and effectiveness is compared which leads to the development of a successful web based applications. Various reasons which differentiate the traditional software development from web system development have also been worked out. The presences of specific features recommended in proposed framework are also tested by available methodologies. Finally, an initial framework is proposed which caters to the need of specific web development process model.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
Similar content being viewed by others

Web Intelligence Linked Open Data for Website Design Reuse

Analysing, Designing, Implementation and Coding E-Commerce Through UX

Application-Driven Approach for Web Site Development
Fitzgerald B, Wojtkowski W, Wrycza S, Zupancic J (1997) System development methods for the next century. Plenum Press, New York
Google Scholar
Escalona MJ, Koch N (2004) Requirements engineering for web applications—A Comparative Study. J Web Eng 2(3):193–212 (© Rinton Press)
Lal M (2011) Web 3.0 in education and research. BVICAM’s Int J Inf Technol 3(2):2–9
Jain R et al (2013) Online rule generation software process model. BIJIT-BVICAM’s Int J Inf Technol 5(1):505–511
Shaffi AS, Al-Obaidy M (2013)Analysis and Comparative Study of traditional and web information systems development methodology (WISDM) towards web development applications. Int J Emerg Technol Adv Eng Website: www.ijetae.com (ISSN 2250-2459, ISO 9001:2008 Certified Journal) 3(11)
Aggarwal N, Soni R (2013) Comparative study of requirement engineering methods. Int J Adv Res Comput Sci Softw Eng 3(7) ISSN: 2277 128X. www.ijarcsse.com
Murugesan S, Deshpande Y, Hansen S, Ginige A (2016) Web engineering: a new discipline for development of web-based systems. Web Eng Lect Notes Comput Sci 2001:3–13
MATH Google Scholar
Omaima NA (2013) Hybrid web engineering process model for the development of large scale web applications. J Theor Appl Inf Technol 53(1):1–5
Clutterbuck P, Rowlands T, Seamons O (2009) A case study of SME web application development effectiveness via agile methods. Electron J Inf Syst Eval 12(1):13–26
Goi CL (2012) The impact of website development on organisation performance: Malaysia’s perspective. Afr J Bus Manage 6(7):2435–2448
Ruhe M, Siemens AG, Jeffery R, Wieczorek I (2003) Using web objects for estimating software development effort for web applications. Software Metrics Symposium, Proceedings. Ninth International, 1530-1435, 0-7695-1987-3, p 30–37
Murugesan S (2008) Web application development: challenges and role of web engineering. Hum Comput Interact Ser 1:7–32. doi: 10.1007/978-1-84628-923-1_2
Article Google Scholar
Watkins C, Marenka S (1995) The internet edge in business. AP Professional, Massachusetts
Manhas J (2015) Comparative Study of cross browser compatibility as design issue in various websites. BVICAM’s Int J Inf Technol 7(1):1–3
Kumar TV, Guruprasad HS (2015) Clustering of web usage data using hybrid K-means and PACT Algorithms. Our Major Index Int Level 4852:871
Download references
Author information
Authors and affiliations.
Department of Computer Sciences and IT (BC), University of Jammu, Jammu, Jammu and kashmir, India
Jatinder Manhas
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Jatinder Manhas .
Rights and permissions
Reprints and permissions
About this article
Manhas, J. Initial framework for website design and development. Int. j. inf. tecnol. 9 , 363–375 (2017). https://doi.org/10.1007/s41870-017-0045-4
Download citation
Received : 05 March 2017
Accepted : 25 September 2017
Published : 23 October 2017
Issue Date : December 2017
DOI : https://doi.org/10.1007/s41870-017-0045-4
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Development
- Methodology
- Find a journal
- Publish with us
- Track your research
The Design and Implementation of Responsive Web Page Based on HTML5 and CSS3
Ieee account.
- Change Username/Password
- Update Address
Purchase Details
- Payment Options
- Order History
- View Purchased Documents
Profile Information
- Communications Preferences
- Profession and Education
- Technical Interests
- US & Canada: +1 800 678 4333
- Worldwide: +1 732 981 0060
- Contact & Support
- About IEEE Xplore
- Accessibility
- Terms of Use
- Nondiscrimination Policy
- Privacy & Opting Out of Cookies
A not-for-profit organization, IEEE is the world's largest technical professional organization dedicated to advancing technology for the benefit of humanity. © Copyright 2024 IEEE - All rights reserved. Use of this web site signifies your agreement to the terms and conditions.
Academia.edu no longer supports Internet Explorer.
To browse Academia.edu and the wider internet faster and more securely, please take a few seconds to upgrade your browser .
Enter the email address you signed up with and we'll email you a reset link.
- We're Hiring!
- Help Center

A Critical Review of Research on Website Development and Design Related to Diversity Issues

Related Papers
Journal of Computer Science IJCSIS , Dr. Mahdi Miraz , Maaruf Ali
Due to mass global migration and increased usage of the Internet, it is now very important to address the cultural aspects of the usability problems of any Information and Communication Technology (ICT) products such as software, websites or applications (apps) whether to be used on PCs, Laptops, Smartphones, Tablets, Smart TVs or any other devices. To augment the " Design for All " concept, this research demonstrates the need to cater for culturally diverse users while designing user interfaces. This has been achieved, by investigating ICT products and conducting an extensive literature survey. The study concludes that it is very important to work on cross-cultural usability problems and bring these issues under focus.
Jennifer Mankoff
ABSTRACT The World Wide Web plays an important role in our society—enabling broader access to information and services than was previously available. However, Website usability and accessibility are still a problem. Numerous, automated, evaluation and transformation tools are available to help Web developers build better sites for users with diverse needs. A survey of these automated tools is presented in the context of the user abilities they support.
Jatinder Manhas
The web is one of the most revolutionary and global technique influencing every aspect of the people throughout the world including culture. The web has become the most intelligible technique throughout the world which enables distribution of services through its websites. The professionals and designers suggest that a well designed user interface is more influential and improves the performance of the website. In this paper, the authors have examined and found out the cultural influences on website design. Culture can be defined as a group of people having certain things in common which could affect the website design. The authors in this paper took different charity websites from North and South India and examined them by comparing the differences in the design with respect to culture. Authors examined these websites for 5 different parameters i.e. Information, Visual design, page layout, user input and multimedia. Findings suggest that north Indian and south Indian websites diffe...
Proceedings of CHI 2001
Carina de Villiers
Shohreh Hadian
The 40th ACM International Conference on Design of Communication
Meghalee Das
Ann Borda , Giuliano Gaia
Iosif Klironomos
International Journal of People-Oriented Programming
Renate Motschnig
Human-Centered Design focuses on the analysis, specification and involvement of a product's end users as driving elements in the design process. The primary research objective of the case-study presented in this paper is to illustrate that it is essential to include users with special needs into all major steps of designing a web-portal that provides services to these special users. But how can this be accomplished in the case of users with special cognitive and affective needs? Would the “classical” Human-Centered Design Process (HCD) be sufficient or would it need to be adapted and complemented with special procedures and tools? In this paper the design team shares the strategies they adopted and the experiences they gained by including users with dyslexia in the design of the LITERACY Web-Portal. Besides providing insight into the special effort and steps needed to adapt HCD for users with special needs, the paper encourages application designers to include end-users even tho...
Achmad Syarief
RELATED PAPERS
Boletim IG-USP. Publicação Especial
Ivone Ksonoki
Luis Aguirre
IEE Proceedings - Radar, Sonar and Navigation
s.koteswara rao
heri nurdiansyah
JOURNAL OF ENGINEERING SCIENCE AND TECHNOLOGY REVIEW
shadia tewfik
George Graen
Jurnal Kelautan Tropis
NIKE IKA NUZULA
Arif Setiawan Kusuma
Jean-Pierre Rey
InterCambios: Dilemas y Transiciones de la Educación Superior
alejandra gatica
Journal of University of Shanghai for Science and Technology
Nor Mashitah Mohd Radzi
Elisa Crisci
Discrete & Computational Geometry
Saugata Basu
The American journal of clinical nutrition
Aoife Caffrey
Molecular Ecology
Carlos Murga Herrera
Proc. Biol. Soc. Wash
Martha Rocha
Hemant K Tiwari
“O‘zbekiston Milliy universitetining ilm-fan rivoji va jamiyat taraqqiyotida tutgan o‘rni” mavzusidagi xalqaro ilmiy-amaliy konferensiya, 2023 yil, 12 may
Dostonbek Kamolov
Journal of Cluster Science
Harsh Sharma
Johan Manuel Mejia Moreno
Pathology oncology research : POR
Chinmay Panda
Animale male
Neiva Guedes
Buy Verified Cash App Accounts
RELATED TOPICS
- We're Hiring!
- Help Center
- Find new research papers in:
- Health Sciences
- Earth Sciences
- Cognitive Science
- Mathematics
- Computer Science
- Academia ©2024
Content Search
Terms of Reference for Terms of Reference (ToR) for White/Research Paper on AI Usage in Humanitarian and Development Sectors for NGOs
Introduction to Required Services
Oxfam has been in Jordan since the 1990s. We work with local partners in Jordan for a future where everyone can reach their full potential, especially women and young people. We provide essential humanitarian assistance to Syrian refugees as well as vulnerable Jordanians, while promoting longer term sustainable solutions to challenges facing Jordan, with a focus on Climate Justice, Economic Justice and Gender Justice.
Oxfam is seeking a consultant(s) to assist in developing a White/Research Paper on AI Usage in Humanitarian and Development Sectors, with a particular emphasis on data empowerment and the creation of an empowered data environment and ecosystem. Additionally, this paper will lay the groundwork for the intentional use of AI, identifying uses across the two sectors, considering staff development needs, and ensuring both risks and opportunities are understood and managed in the process.
The main purpose of this paper is to investigate the current and potential applications and impact of AI in humanitarian and development sectors. It will also provide guidance and recommendations for Oxfam and other stakeholders on how to use AI responsibly, effectively, and ethically, with a focus on data empowerment.
Scope of SERVICES
- Conduct a comprehensive literature review and stakeholder analysis.
- Identify and analyze case studies showcasing successful AI implementations in humanitarian and development initiatives worldwide.
- Identify and analyze the opportunities and challenges of using AI in humanitarian and development contexts, with a focus on data empowerment.
- Develop a framework and a set of principles for the intentional use of AI in humanitarian and development sectors.
- Provide recommendations and action points on how to integrate AI in strategies, policies, and operations to leverage AI for humanitarian and development goals.
Suggested Research Questions
- What are the existing trends and patterns of AI usage in humanitarian and development sectors? What are the main drivers and barriers for its adoption and/or scaling? How does data empowerment factor into these trends?
- What are the potential benefits and risks of using AI in development and humanitarian sectors, and how can these be measured and mitigated?
- What are the existing and needed frameworks and principles to enable the responsible and ethical use of AI, and what are the ethical, legal, and social implications of using AI in the development and humanitarian sectors?
- How can NGOs and other stakeholders use AI to improve their humanitarian and development outcomes? What are best practices and lessons learned from existing AI projects and initiatives?
- What are the gaps and needs in terms of capacities, skills, resources, and partnerships for NGOs and other stakeholders to effectively and sustainably use AI in humanitarian and development sectors, and how can they be addressed?
*Note: These are suggested research questions its up to the consultant after consultations with Oxfam team to develop research questions.
Deliverables & TIMEFRAME
Description
# Of Days Required
Inception Report and Plan
Develop an inception report outlining objectives, methodologies, and timelines. Establish a detailed work plan for project execution.
Literature review and stakeholder analysis
Reviewing existing sources and conducting interviews or surveys with relevant stakeholders to understand the current state of AI usage in humanitarian and development sectors.
Case studies analysis
Identifying and analyzing examples of successful AI projects and initiatives in humanitarian and development contexts, highlighting the impact, the challenges, and the lessons learned.
Opportunities and challenges analysis
Identifying and analyzing the potential benefits and risks of using AI in humanitarian and development contexts, and the factors that influence them.
Framework and principles development
Developing a framework and a set of principles for the intentional use of AI in humanitarian and development sectors.
Recommendations and action points
Providing recommendations and action points for Oxfam and other stakeholders on how to integrate AI into their strategies, policies, and operations, and how to build their capacities and partnerships to leverage AI for humanitarian and development goals.
Required Qualifications
The research entity submitting a proposal for this case study should possess the following qualifications:
- Experience in project management, preferably in research or consultancy projects related to humanitarian aid, development, or technology.
- Strong research skills, including proficiency in conducting literature reviews, synthesizing information, and analyzing data.
- Familiarity with AI applications and technologies, particularly in the context of humanitarian and development sectors.
- Knowledge of ethical considerations surrounding AI deployment, including issues related to privacy, bias, transparency, and accountability.
- Excellent writing and communication skills, with the ability to draft clear, concise, and well-structured reports or papers.
- Experience in stakeholder engagement and facilitation, including conducting interviews, focus groups, or workshops with diverse groups.
- Ability to work independently, meet deadlines, and manage multiple tasks simultaneously.
- Understanding of gender and diversity considerations in humanitarian and development programming is desirable.
Top of Form
Evaluation Criteria
Applications will be assessed according to the following criteria:
- Financial proposal 30%: reasonable budget for the proposed goals and activities and proper budget breakdown.
- Qualifications and competencies 20%: relevant experience of the consultant who will be conducting the assignment in Jordan
- Technical proposal 50%: Completeness of the proposal including: the approach, the methodology, Timeline, expected output(s).
How to apply
Submission Requirements
Please submit your application to: [email protected], including each of the following separately, and valid for 90 days:
- Covering letter
- Technical offer explaining the methodology and approach
- Financial offer including all costs, in Jordanian Dinars (JOD), inclusive 5% income tax for Jordanians, and 10 % income tax for internationals.
GUIDANCE ON HOW TO APPLY
Applicants should submit technical and financial proposals as separate documents to [email protected] with the subject line: [INSERT TITLE] and RFQ Number [RFQ-JOAMM-24-0047].
Related Content
Jordan + 2 more
WFP Jordan Country Brief, March 2024
Complementary pathways jordan, january - march 2024.
Jordan + 1 more
Jordan: Communication with Communities Dashboard (March 2024)
Jordan: resettlement dashboard | march 2024.

AI + Machine Learning , Announcements , Azure AI , Azure AI Studio
Introducing Phi-3: Redefining what’s possible with SLMs
By Misha Bilenko Corporate Vice President, Microsoft GenAI
Posted on April 23, 2024 4 min read
- Tag: Copilot
- Tag: Generative AI
We are excited to introduce Phi-3, a family of open AI models developed by Microsoft. Phi-3 models are the most capable and cost-effective small language models (SLMs) available, outperforming models of the same size and next size up across a variety of language, reasoning, coding, and math benchmarks. This release expands the selection of high-quality models for customers, offering more practical choices as they compose and build generative AI applications.
Starting today, Phi-3-mini , a 3.8B language model is available on Microsoft Azure AI Studio , Hugging Face , and Ollama .
- Phi-3-mini is available in two context-length variants—4K and 128K tokens. It is the first model in its class to support a context window of up to 128K tokens, with little impact on quality.
- It is instruction-tuned, meaning that it’s trained to follow different types of instructions reflecting how people normally communicate. This ensures the model is ready to use out-of-the-box.
- It is available on Azure AI to take advantage of the deploy-eval-finetune toolchain, and is available on Ollama for developers to run locally on their laptops.
- It has been optimized for ONNX Runtime with support for Windows DirectML along with cross-platform support across graphics processing unit (GPU), CPU, and even mobile hardware.
- It is also available as an NVIDIA NIM microservice with a standard API interface that can be deployed anywhere. And has been optimized for NVIDIA GPUs .
In the coming weeks, additional models will be added to Phi-3 family to offer customers even more flexibility across the quality-cost curve. Phi-3-small (7B) and Phi-3-medium (14B) will be available in the Azure AI model catalog and other model gardens shortly.
Microsoft continues to offer the best models across the quality-cost curve and today’s Phi-3 release expands the selection of models with state-of-the-art small models.

Azure AI Studio
Phi-3-mini is now available
Groundbreaking performance at a small size
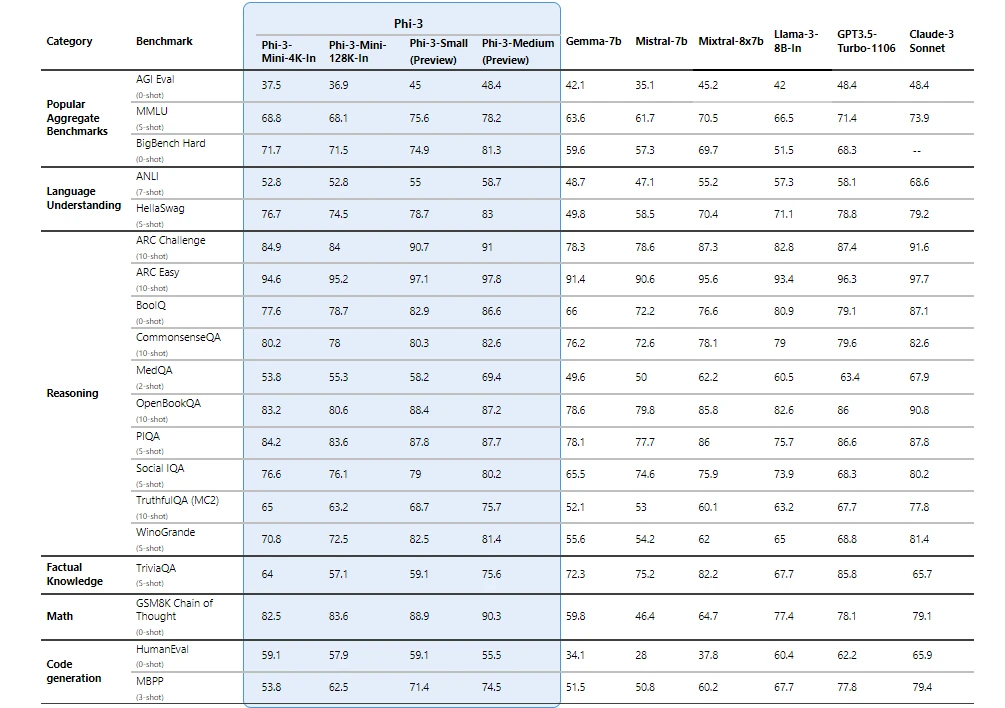
Phi-3 models significantly outperform language models of the same and larger sizes on key benchmarks (see benchmark numbers below, higher is better). Phi-3-mini does better than models twice its size, and Phi-3-small and Phi-3-medium outperform much larger models, including GPT-3.5T.
All reported numbers are produced with the same pipeline to ensure that the numbers are comparable. As a result, these numbers may differ from other published numbers due to slight differences in the evaluation methodology. More details on benchmarks are provided in our technical paper .
Note: Phi-3 models do not perform as well on factual knowledge benchmarks (such as TriviaQA) as the smaller model size results in less capacity to retain facts.
Safety-first model design
Responsible ai principles
Phi-3 models were developed in accordance with the Microsoft Responsible AI Standard , which is a company-wide set of requirements based on the following six principles: accountability, transparency, fairness, reliability and safety, privacy and security, and inclusiveness. Phi-3 models underwent rigorous safety measurement and evaluation, red-teaming, sensitive use review, and adherence to security guidance to help ensure that these models are responsibly developed, tested, and deployed in alignment with Microsoft’s standards and best practices.
Building on our prior work with Phi models (“ Textbooks Are All You Need ”), Phi-3 models are also trained using high-quality data. They were further improved with extensive safety post-training, including reinforcement learning from human feedback (RLHF), automated testing and evaluations across dozens of harm categories, and manual red-teaming. Our approach to safety training and evaluations are detailed in our technical paper , and we outline recommended uses and limitations in the model cards. See the model card collection .
Unlocking new capabilities
Microsoft’s experience shipping copilots and enabling customers to transform their businesses with generative AI using Azure AI has highlighted the growing need for different-size models across the quality-cost curve for different tasks. Small language models, like Phi-3, are especially great for:
- Resource constrained environments including on-device and offline inference scenarios.
- Latency bound scenarios where fast response times are critical.
- Cost constrained use cases, particularly those with simpler tasks.
For more on small language models, see our Microsoft Source Blog .
Thanks to their smaller size, Phi-3 models can be used in compute-limited inference environments. Phi-3-mini, in particular, can be used on-device, especially when further optimized with ONNX Runtime for cross-platform availability. The smaller size of Phi-3 models also makes fine-tuning or customization easier and more affordable. In addition, their lower computational needs make them a lower cost option with much better latency. The longer context window enables taking in and reasoning over large text content—documents, web pages, code, and more. Phi-3-mini demonstrates strong reasoning and logic capabilities, making it a good candidate for analytical tasks.
Customers are already building solutions with Phi-3. One example where Phi-3 is already demonstrating value is in agriculture, where internet might not be readily accessible. Powerful small models like Phi-3 along with Microsoft copilot templates are available to farmers at the point of need and provide the additional benefit of running at reduced cost, making AI technologies even more accessible.
ITC, a leading business conglomerate based in India, is leveraging Phi-3 as part of their continued collaboration with Microsoft on the copilot for Krishi Mitra, a farmer-facing app that reaches over a million farmers.
“ Our goal with the Krishi Mitra copilot is to improve efficiency while maintaining the accuracy of a large language model. We are excited to partner with Microsoft on using fine-tuned versions of Phi-3 to meet both our goals—efficiency and accuracy! ” Saif Naik, Head of Technology, ITCMAARS
Originating in Microsoft Research, Phi models have been broadly used, with Phi-2 downloaded over 2 million times. The Phi series of models have achieved remarkable performance with strategic data curation and innovative scaling. Starting with Phi-1, a model used for Python coding, to Phi-1.5, enhancing reasoning and understanding, and then to Phi-2, a 2.7 billion-parameter model outperforming those up to 25 times its size in language comprehension. 1 Each iteration has leveraged high-quality training data and knowledge transfer techniques to challenge conventional scaling laws.
Get started today
To experience Phi-3 for yourself, start with playing with the model on Azure AI Playground . You can also find the model on the Hugging Chat playground . Start building with and customizing Phi-3 for your scenarios using the Azure AI Studio . Join us to learn more about Phi-3 during a special live stream of the AI Show.
1 Microsoft Research Blog, Phi-2: The surprising power of small language models, December 12, 2023 .
Let us know what you think of Azure and what you would like to see in the future.
Provide feedback
Build your cloud computing and Azure skills with free courses by Microsoft Learn.
Explore Azure learning
Related posts
AI + Machine Learning , Azure AI , Azure AI Content Safety , Azure Cognitive Search , Azure Kubernetes Service (AKS) , Azure OpenAI Service , Customer stories
AI-powered dialogues: Global telecommunications with Azure OpenAI Service chevron_right
AI + Machine Learning , Azure AI , Azure AI Content Safety , Azure OpenAI Service , Customer stories
Generative AI and the path to personalized medicine with Microsoft Azure chevron_right
AI + Machine Learning , Azure AI , Azure AI Services , Azure AI Studio , Azure OpenAI Service , Best practices
AI study guide: The no-cost tools from Microsoft to jump start your generative AI journey chevron_right
AI + Machine Learning , Azure AI , Azure VMware Solution , Events , Microsoft Copilot for Azure , Microsoft Defender for Cloud
Get ready for AI at the Migrate to Innovate digital event chevron_right
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 24 April 2024
Spatiotemporally resolved colorectal oncogenesis in mini-colons ex vivo
- L. Francisco Lorenzo-Martín ORCID: orcid.org/0000-0003-4717-9338 1 na1 ,
- Tania Hübscher ORCID: orcid.org/0000-0002-2376-712X 1 na1 ,
- Amber D. Bowler ORCID: orcid.org/0000-0002-9439-2839 2 , 3 ,
- Nicolas Broguiere ORCID: orcid.org/0000-0001-9934-4505 1 ,
- Jakob Langer ORCID: orcid.org/0000-0002-0095-1936 1 ,
- Lucie Tillard 1 ,
- Mikhail Nikolaev ORCID: orcid.org/0000-0001-5955-900X 4 ,
- Freddy Radtke ORCID: orcid.org/0000-0003-4315-4045 2 , 3 &
- Matthias P. Lutolf ORCID: orcid.org/0000-0002-5898-305X 1 , 4
Nature ( 2024 ) Cite this article
15k Accesses
1 Citations
389 Altmetric
Metrics details
- Cancer models
- Gastrointestinal cancer
- Lab-on-a-chip
- Tissue engineering
Three-dimensional organoid culture technologies have revolutionized cancer research by allowing for more realistic and scalable reproductions of both tumour and microenvironmental structures 1 , 2 , 3 . This has enabled better modelling of low-complexity cancer cell behaviours that occur over relatively short periods of time 4 . However, available organoid systems do not capture the intricate evolutionary process of cancer development in terms of tissue architecture, cell diversity, homeostasis and lifespan. As a consequence, oncogenesis and tumour formation studies are not possible in vitro and instead require the extensive use of animal models, which provide limited spatiotemporal resolution of cellular dynamics and come at a considerable cost in terms of resources and animal lives. Here we developed topobiologically complex mini-colons that are able to undergo tumorigenesis ex vivo by integrating microfabrication, optogenetic and tissue engineering approaches. With this system, tumorigenic transformation can be spatiotemporally controlled by directing oncogenic activation through blue-light exposure, and emergent colon tumours can be tracked in real-time at the single-cell resolution for several weeks without breaking the culture. These induced mini-colons display rich intratumoural and intertumoural diversity and recapitulate key pathophysiological hallmarks displayed by colorectal tumours in vivo. By fine-tuning cell-intrinsic and cell-extrinsic parameters, mini-colons can be used to identify tumorigenic determinants and pharmacological opportunities. As a whole, our study paves the way for cancer initiation research outside living organisms.
Similar content being viewed by others

High-throughput automated organoid culture via stem-cell aggregation in microcavity arrays
Three-dimensional culture models mimic colon cancer heterogeneity induced by different microenvironments
Engineering organoids
Cancer arises through the accumulation of genetic lesions that confer unrestrained cell growth potential. Over the past 70 years, both two-dimensional (2D) and three-dimensional (3D) in vitro culture models have been developed to make simplified, animal-free versions of cancers readily available for research 4 . These models successfully portray and dissect a wide range of relatively simple cancer cell behaviours, such as proliferation, motility, invasiveness, survival, cell–cell and cell–stroma interactions, and drug responses, among others 1 , 2 , 4 . However, modelling more complex processes that involve multiple cell (sub)types and tissue-level organization remains a challenge, as is the case for cancer initiation.
The cellular transition from healthy to cancerous is an intricate evolutionary process that is still largely obscure due to the insufficient topobiological complexity of the available in vitro cell culture systems, which precludes de novo tumour generation and the establishment of pathophysiologically relevant tumorigenic models 5 , 6 . Even the current gold-standard organoid-based 3D models, which are often postulated as a bridge between in vitro and in vivo 1 , 3 , 7 , are too simplified for modelling cancer development ex vivo. This is mostly due to (1) their closed cystic structure instead of an in vivo-like apically open architecture 8 ; (2) their short lifespan that requires breaking up the culture every few days for passaging 9 ; (3) their lack of topobiological stability and consistency owing to their stochastic growth in 3D matrices 8 ; and (4) their inability to generate hybrid tissues composed of healthy and cancer cells in a balanced and integrated manner 10 . Various next-generation approaches such as bioprinting and microfabrication technologies have been recently implemented to partially address some of these issues 11 , 12 ; however, none have been able to fully recreate intratumour and intertumour complexity. Consequently, cancer research is still inevitably bound to animal experimentation, which provides a pathophysiologically relevant setting, but forbids high-resolution and real-time analyses of cellular dynamics during oncogenesis. Moreover, these models are economically and ethically costly. Thus, while there is the widespread consensus that animal use in research should be reduced, replaced and refined (the 3 Rs 13 ), this commitment is severely hindered by the insufficient physiological complexity displayed by classical in vitro systems.
Here we postulated that a 3D system able to solve the existing limitations of in vitro cultures could be engineered by leveraging scaffold-guided organoid morphogenesis and optogenetics. Specifically, we developed miniature colon tissues in which cells could (1) be cultured for long durations (several weeks) without the need for breaking the culture through passaging; (2) reproduce the stem-differentiated cell patterning axis in a stable and anatomically relevant topology; (3) be easily mutated and tracked in a spatiotemporally controlled manner; and (4) create a biomechanically dynamic system that allows for tumour emergence while preserving the integrity of the surrounding healthy tissue. These features permit the development of biologically complex tumours ex vivo, bridging the gap between in vitro and in vivo models by providing a high-resolution system that can be used to dissect the molecular factors orchestrating cancer initiation.
Spatiotemporally regulated tumorigenesis
We focused on colorectal cancer (CRC) as it is one of the most prominent cancer types worldwide and its malignant transformation can be readily engineered genetically 14 , 15 . To first achieve spatiotemporal control of oncogenic DNA recombination, we developed a doxycycline-sensitive blue-light-regulated Cre system (hereafter, OptoCre), which we then introduced into inducible Apc fl/fl Kras LSL-G12D/+ Trp53 fl/fl (AKP) healthy colon organoids (Extended Data Fig. 1a–c ). A fluorescent Cre reporter was also incorporated to track cells that undergo oncogenic recombination (Extended Data Fig. 1b,c ). We initially tested the system in conventional organoid cultures, in which OptoCre efficiently induced recombination in the presence of blue light and doxycycline (Extended Data Fig. 1d,e ). Dosage optimization prevented unwanted activation by coupling high efficiency with low leakiness (~1.6%) (Extended Data Fig. 1d,e ). To confirm successful oncogenic transformation, we removed growth factors (EGF, noggin, R-spondin, WNT3A) from the organoid medium and observed that only cells with an activated OptoCre were able to grow, a well-known hallmark of mutated AKP colon organoids 16 (Extended Data Fig. 1f ). The presence of the expected mutations at the Apc, Kras and Trp53 loci was confirmed by PCR and exome sequencing (see below; Extended Data Fig. 3f,g ).
On the basis of previous evidence that small intestine cells can form stable tube-shaped epithelia through scaffold-guided organoid morphogenesis in microfluidic devices 9 , we next aimed to establish a ‘mini-colon’ constituted by OptoCre-AKP cells. By seeding colon cell suspensions in hydrogel-patterned microfluidic devices, we generated single-layered colonic epithelia spatially arranged into crypt- and lumen-like domains (Extended Data Fig. 2a ). This spatial arrangement recapitulated the spatial distribution found in vivo, with stem and progenitor (SOX9 + ) cells located at the bottom of the crypt domains and more differentiated colonocytes (FABP1 + ) located in the upper crypt and lumen areas 17 , 18 (Extended Data Fig. 2b ). In contrast to conventional colon organoids, the lumen of these mini-colons was readily perfusable with fresh medium, enabling the removal of cell debris and extending their lifespan to several weeks without the need for passaging or tissue disruption (Extended Data Fig. 2a ).
Once the healthy mini-colon system was established, we investigated its potential to capture tumour biology by inducing oncogenic recombination through blue-light illumination (Fig. 1a ). To mimic the scenario found in vivo, we fine-tuned OptoCre activation to mutate only a small number of cells (<0.5% of the total population). Due to the stability and defined topology of the mini-colon, we easily detected the acquisition of AKP mutations at the single-cell level (GFP + cells) and tracked their evolution over time (Extended Data Fig. 2c,d ). This revealed that cell death is one of the earliest responses to oncogenic recombination, as mutated mini-colons displayed higher cell shedding rates compared with the controls (Extended Data Fig. 2e ), with a large fraction of the mutated cells undergoing apoptosis (Supplementary Video 1 ). Nevertheless, some mutated cells escaped apoptosis and, after a quiescent period (24–72 h), started dividing at an accelerated pace (Extended Data Fig. 2d ). In conventional organoid cultures, these fast-proliferating mutated cells did not lead to any overt tissular rearrangements (Fig. 1b ), whereas, in the mini-colon system, they developed neoplastic structures over 5–10 days (Fig. 1b ). Furthermore, these mini-colon neoplasias evolved from polyp-like to full-blown tumours, recapitulating in vivo tumorigenesis (Fig. 1b,c and Supplementary Videos 2 and 3 ).
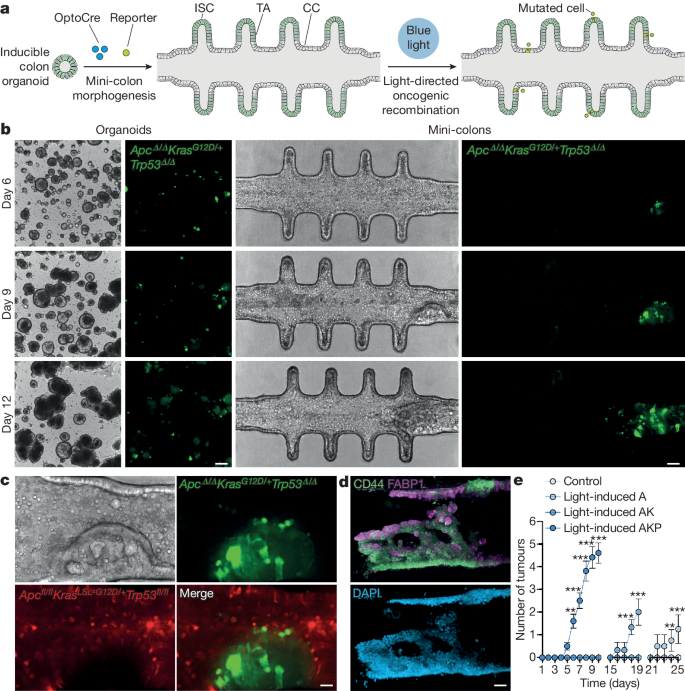
a , Schematic of the experimental workflow followed to induce tumorigenesis in mini-colons. CC, colonocyte; ISC, intestinal stem cell; TA, transit-amplifying cell. b , Bright-field and fluorescence images of time-course tumorigenesis experiments in conventional organoids and mini-colons. Fluorescence signal indicates oncogenic recombination. Scale bars, 200 μm (left) and 75 μm (right). c , Bright-field and fluorescence close-up images of a mini-colon tumour. The red and green signals correspond to healthy and mutated cells, respectively. Scale bar, 25 μm. d , Immunofluorescence images of a mini-colon tumour showing the presence of CD44 (top, green), FABP1 (top, magenta) and nuclei (bottom). Scale bar, 35 μm. e , Multiplicity of tumours emerged in mini-colons of the indicated genotypes after light-mediated oncogenic induction. Statistical analysis was performed using two-way analysis of variance (ANOVA) with Sidak’s multiple-comparison test; ** P = 0.024 (day 6, AKP), ** P = 0.0021 (day 24, A), *** P < 0.0001 (all other conditions). n = 5, 4, 3 and 10 mini-colons for the control, light-induced A, light-induced AK and light-induced AKP conditions, respectively. Data are mean ± s.e.m.
Source Data
Immunostaining analyses revealed that these tumours stemmed from CD44 high cells—a bona fide marker for cancer stem cells in vivo 19 —at the base of the epithelium (Fig. 1d , Extended Data Fig. 2f and Supplementary Video 4 ). Conversely, the bulk of the tumours was composed of cells with different degrees of differentiation, as revealed by the downregulation and upregulation of CD44 and FABP1, respectively (Fig. 1d and Supplementary Video 5 ). This indicated the existence of intratumour heterogeneity in the mini-colon, resembling the in vivo scenario 20 . Consistent with this, histopathological studies showed that these tumours displayed the histological organization characteristic of tubular adenomas (Extended Data Fig. 3a ). To validate their cancerous nature, we performed transplantation experiments in immunodeficient mice and found that mini-colon-derived cancer cells formed tumours in vivo with undistinguishable efficiency from bona fide tumour-derived cancer cells (Extended Data Fig. 3b,c ). Moreover, their histopathological structure was also comparable to the one displayed by primary tumours developed in the colon of AKP mice (Extended Data Fig. 3d ) and included the presence of locally invasive nodules and areas with adenocarcinoma-like features (Extended Data Fig. 3e ).
We confirmed through PCR and exome sequencing that tumour development in the mini-colon was directly associated with the expected mutations at the Apc, Kras and Trp53 loci (Extended Data Fig. 3f,g ). Consistent with this, using organoid lines with a reduced mutational burden ( Apc fl/fl Kras LSL-G12D/+ (hereafter, AK) and Apc fl/fl (hereafter, A)) produced longer latencies in tumour development in a dosage-dependent manner (Fig. 1e and Extended Data Fig. 3h ), demonstrating that mini-colon tumorigenesis can be modulated by the number of oncogenic driver mutations. Collectively, these data show that the mini-colon system enables spatiotemporally controlled in vitro modelling of CRC tumorigenesis with a considerable degree of topobiological complexity.
Context-dependent tumorigenic plasticity
Careful examination of induced mini-colons revealed consistent morphological differences among tumours according to their initiation site, with prominent dense or cystic internal structures arising from the crypt and the luminal epithelium, respectively (see below; Fig. 2b (top)). As mini-colons comprise different types of cells along the crypt–lumen axis (Extended Data Fig. 2b ), we leveraged the spatial resolution provided by OptoCre to investigate whether the initiating cell niche conditioned the morphological and functional features of nascent tumours. To spatially control AKP mutagenesis, we coupled the mini-colon to a photomask restricting blue-light exposure to specific regions of the colonic epithelium (Fig. 2a ), which provided low off-target recombination rates (around 8.5%) (Fig. 2b,c and Extended Data Fig. 4a ). Here again, dense and cystic tumours developed when crypt and lumen epithelia, respectively, were mutationally targeted by blue light (Fig. 2b ). To confirm that this was associated with the differentiation status of the tumour-initiating cell, we cultured mini-colons in either low- or high-differentiation medium before oncogenic induction to shift the proportions of (un)differentiated cells. Low-differentiation conditions produced mini-colons with thicker epithelia, early tumour development and a reduced fraction of cystic tumours (Fig. 2d and Extended Data Fig. 4b,c ). Conversely, high-differentiation conditions produced mini-colons with thinner epithelia, delayed tumour formation and increased cystic tumour frequency (Fig. 2d and Extended Data Fig. 4b,c ). These results indicate that the different environments of the mini-colon can shape tumour fate.
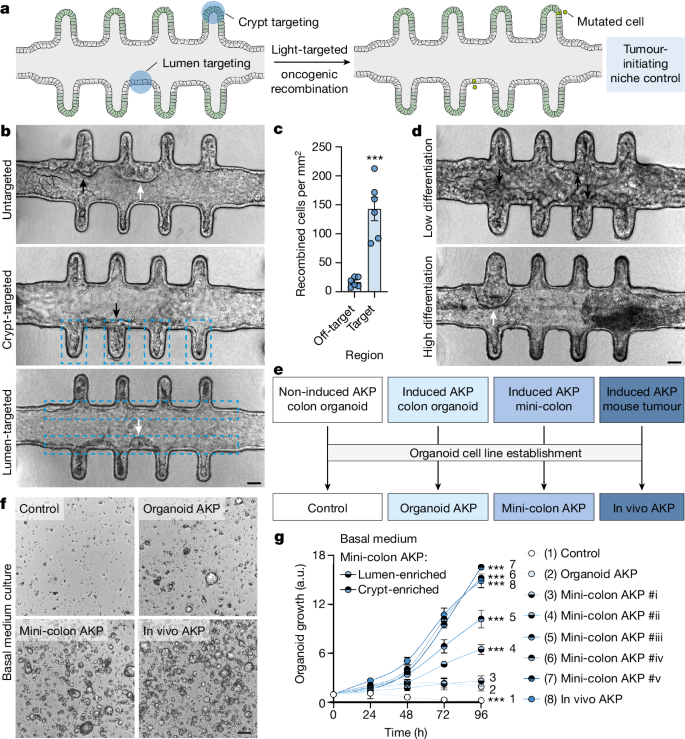
a , Schematic of the experimental workflow followed to spatiotemporally target tumorigenesis in mini-colons. b , Bright-field images of mini-colons that have undergone untargeted (top), crypt-targeted (middle) and lumen-targeted (bottom) tumorigenesis. Targeted areas are indicated by dashed blue lines. The black and white arrows indicate tumours with compact and cystic morphologies, respectively. Scale bar, 75 μm. c , The oncogenic recombination efficiency in targeted and off-target areas in mini-colons. Statistical analysis was performed using two-tailed t -tests; *** P < 0.0001. n = 6 mini-colons per condition. Each point represents one mini-colon. d , Bright-field images of induced mini-colons cultured in low-differentiation (top, WENRNi) and high-differentiation (bottom, ENR) conditions. The black and white arrows indicate tumours with compact and cystic morphologies, respectively. Scale bar, 75 μm. e , Schematic of the different colon organoid lines generated in this work. f , Bright-field images of the indicated colon organoid lines cultured for 2 days in basal medium. Scale bar, 200 μm. g , Metabolic activity (measured using resazurin) of the indicated colon organoid lines cultured in basal medium for the indicated time. Numerical labelling (1–8) was used to facilitate cell line identification. Statistical analysis was performed using two-way ANOVA with Sidak’s multiple-comparison test; *** P = 0.0004 (control), *** P < 0.0001 (all other conditions). n = 3 cultures for each line. For c and g , data are mean ± s.e.m.
To evaluate the functional repercussions of the tumour-initiating niche, we isolated cancer cells from mini-colons enriched in either crypt- or lumen-derived tumours and established organoid cell lines (termed mini-colon AKP) (Fig. 2e ). As a control, we generated AKP mutant organoids by shining blue light onto inducible organoids and kept these mutants in parallel with their mini-colon equivalents, doing the required passages on confluency (termed organoid AKP) (Fig. 2e ). We also established organoid cultures from AKP colon tumours extracted from tamoxifen-treated Cdx2-cre ERT2 AKP mice (termed in vivo AKP) (Fig. 2e ). Notably, in contrast to mini-colons, none of these three types of mutant AKP lines were morphologically distinguishable from healthy non-mutated cells when cultured as organoids (Fig. 1b and Extended Data Fig. 4d ). When we cultured these organoids in basal medium depleted of growth factors (BM; Methods ), both in vivo and crypt tumour-derived mini-colon AKP organoids preserved their proliferative potential (Fig. 2f,g ). Conversely, organoid and lumen tumour-enriched mini-colon AKP lines displayed significantly reduced proliferation rates (Fig. 2f,g ). This was not due to intrinsic cycling defects in any of the organoid lines tested, as these differences were not observed in standard cancer organoid medium (BMGF; Methods and Extended Data Fig. 4e ). As expected, healthy organoids did not grow in any of these conditions (Fig. 2f,g and Extended Data Fig. 4e ). Collectively, these results show that there are context-dependent factors aside from the founding AKP mutations that condition the growth potential of AKP cells. They also indicate that the cells derived from mini-colon crypt tumours recapitulate the growth properties of in vivo CRC cells more faithfully than conventional organoids.
To investigate the molecular programs underpinning these observations, we profiled the transcriptome of the different AKP lines using RNA sequencing (RNA-seq). We first characterized the differences between the two AKP lines derived from conventional systems, in vivo and organoid AKP cells, which also had the biggest disparity in growth potential (Fig. 2g ). According to our previous experiments, in vivo AKP cells upregulated many genes involved in canonical cancer pathways and the promotion of cell growth (Extended Data Fig. 4f,g ). Conversely, these cells downregulated genes associated with cell differentiation, patterning and transcriptional regulation (Extended Data Fig. 4f,g ). To evaluate whether mini-colon AKP cells recapitulated this in vivo AKP transcriptional signature, we performed single-sample gene set enrichment analysis (GSEA) across all of the cell lines. Here, most of the mini-colon AKP lines outscored their organoid AKP counterparts, especially those derived from crypt tumours (Extended Data Fig. 4h ). To investigate the transcriptional divergence between crypt- and lumen-enriched mini-colon AKP cells, we compared the lines with the highest (#v, crypt-enriched) and lowest (#i, lumen-enriched) in vivo AKP signature score (Extended Data Fig. 4h ). These analyses revealed that crypt-derived mini-colon AKP cells upregulated genes involved in WNT signalling, stem cell pluripotency, lipid metabolism and other pathways involved in cancer (Extended Data Fig. 4i ). To identify the potential drivers of growth factor independence among these, we searched for overlaps between AKP lines with high growth potential in BM (in vivo AKP, mini-colon AKP #v). We found that the latter overexpressed a collection of genes that is involved in the activation of MAPK cascades, including receptor tyrosine kinases (RTKs), G-protein-coupled receptors and soluble factors (Extended Data Fig. 5a ). We therefore theorized that these cells were engaging a surplus of MAPK signalling that gave them a greater fitness under growth-factor-poor conditions. To validate this idea, we tested their response to a panel of inhibitors, which confirmed that the growth of AKP lines in BM heavily relied on signals from RTKs (Extended Data Fig. 5b,c ; regorafenib), including KIT (Extended Data Fig. 5b,c ; ripretinib) and FGF receptors (Extended Data Fig. 5b,c ; infigratinib). Corroborating this, the ligands for these RTKs (SCF, FGF2) and others involved in colonocyte clonogenicity (IGF1) 21 could enhance the growth of the AKP lines with poor proliferation potential in BM (Extended Data Fig. 5d,e ). Importantly, all of these dependencies were either reduced or not detectable in conventional CRC organoid medium (BMGF) (Extended Data Figs. 4e and 5b,c ). Taken together, these data indicate that the mini-colon is a plastic system in which context-dependent factors can drive different functional features in CRC cells, including the engagement of ancillary RTK signals that boost their growth potential in challenging environments.
Intra- and intertumour heterogeneity
We hypothesized that the diversity observed in tumour morphology and growth potential reflected clonally distinct tumour types being initiated in the mini-colon. To validate this idea, we performed single-cell transcriptomic profiling of tumour-bearing mini-colons incorporating a genetic cell barcoding system 22 to preserve clonal information (Fig. 3a ). On the basis of bona fide transcriptional markers, mini-colons comprised eight major cell types that were segregated into undifferentiated, absorptive and secretory lineages (Fig. 3b ). Undifferentiated ( Krt20 − ) cells included stem ( Lgr 5 + ), actively proliferating ( Mki67 + ) and progenitor ( Sox9 + Cd44 + ) cells (Fig. 3b,c and Extended Data Fig. 6a ). Mature ( Krt20 + ) absorptive colonocytes constituted the largest fraction of the mini-colon, and included bottom, middle and top colonocytes based on zonation markers 23 (such as Aldob , Iqgap2 and Clca4a ) (Fig. 3b,c and Extended Data Fig. 6a ). Mucus-producing goblet cells ( Muc2 + ) and hormone-releasing enteroendocrine cells ( Neurod1 + ) constituted the secretory compartment (Fig. 3b,c and Extended Data Fig. 6a ). Collectively, this diverse in vivo-like cell composition indicates that mini-colons provide a physiologically relevant context for conducting oncogenesis studies.
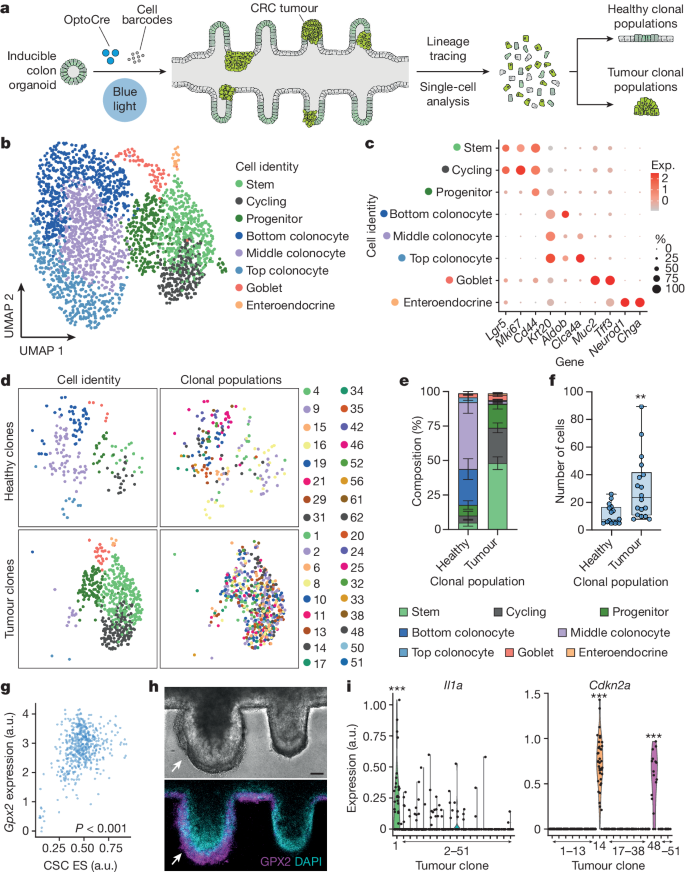
a , Schematic of the experimental workflow followed for single-cell and lineage-tracing analysis of mini-colons. b , Unsupervised uniform manifold approximation and projection (UMAP) clustering of the main cell types in mini-colons 7 days after tumorigenic induction. c , The expression (Exp.) of representative cell-type-specific markers in the different cell populations comprising mini-colons. d , Unsupervised clustering (UMAP) of healthy (top) and tumour (bottom) clonal populations in mini-colons. The cell type (left; colour coded as in b ) and clonal identity (right) are indicated. e , The relative cell type abundance in healthy and tumour mini-colon clonal populations. Data are mean ± s.e.m. n = 16 and 18 for healthy and tumour clones, respectively. f , Healthy and tumour mini-colon clonal population sizes. Statistical analysis was performed using two-tailed Mann–Whitney U -tests; ** P = 0.0011. n = 16 and 18 for healthy and tumour clones, respectively. The box plots show the median (centre lines), the first and third quartiles (box limits) and the minimum and maximum values (whiskers). Each point represents one clonal population. g , The correlation between Gpx2 expression and cancer stem cell transcriptional signature enrichment ( Cd44 , Lgr5 , Sox9 ). Statistical analysis was performed using two-sided Pearson correlation tests; P < 0.0001. n = 540 cells. Each point represents one cell. CSC, cancer stem cell; ES, enrichment score. h , Bright-field and immunofluorescence images showing the abundance of GPX2 (magenta) and nuclei (cyan) in healthy (right) and tumour (left, indicated by arrows) crypts in a mini-colon. Scale bar, 35 μm. i , Expression of the indicated genes in the indicated tumour clones. Statistical analysis was performed using two-sided Wilcoxon rank-sum tests; *** P = 1.77 × 10 −17 ( Il1a , clone 1), 1.00 × 10 −78 ( Cdkn2a , clone 14), 3.67 × 10 −22 ( Cdkn2a , clone 48). n = 540 cells. Each point represents one cell.
To determine the clonal identities across the mini-colon, we compared the genetic barcodes among cells and detected 83 clonal populations. We then discarded small (<5 cells) clones and identified cells containing reads corresponding to the mutated versions of Apc and Trp53 (Extended Data Fig. 6b,c ). These bona fide tumour cells distinguished tumour clonal populations (18 classified) from healthy counterparts (16 classified) ( Methods and Extended Data Fig. 6d ). On average, healthy clonal populations consisted of around 18% undifferentiated cells, which gave rise to the remaining approximately 82% of absorptive colonocytes and secretory cells (Fig. 3d,e ). Conversely, mini-colon tumours were mostly formed by undifferentiated cells (~92%), with sparsely present colonocytes and secretory cells (Fig. 3d,e ). Tumour cells also formed larger clonal populations compared with their healthy counterparts (Fig. 3f ). These cell proportions are well aligned with the ones commonly observed in vivo 24 , 25 .
Analyses of the internal structure of single clonal tumours showed that they comprised a non-homogeneous collection of cells with diverse proliferation, stemness and differentiation markers (Extended Data Fig. 7a ). Such intratumour heterogeneity reflects the complexity of mini-colon tumours, consistent with our immunostaining data (Fig. 1d ). To investigate the mechanisms orchestrating cancer stemness and tumour development, we analysed the transcriptional differences between differentiated ( Krt20 + Apoc2 + Fabp2 + ) and stem ( Lgr5 + Cd44 + Sox9 + ) cancer cells within tumours. We found that Gpx2 , a glutathione peroxidase recently linked to CRC malignant transformation 24 , strongly correlated with the stemness potential of mini-colon cancer cells (Fig. 3g ). Consistent with this, we observed that GPX2 protein was particularly enriched in the basal cells of mini-colon tumours (Fig. 3h ).
To examine whether mini-colons could produce different types of tumours, we next compared the transcriptional profiles of the different tumour clones. Even though all tumour-initiating cells carried the same founding AKP mutations and shared many molecular features, we found clear diversity across mini-colon tumours (Extended Data Fig. 7b ). For example, the expression of the interleukin Il1a and leukocyte peptidase inhibitor Slpi revealed the presence of tumours with an inflammatory-like profile (Fig. 3i and Extended Data Fig. 7b,c ). Cdkn2a (encoding tumour suppressors p14 and p16) and Prdm16 were exclusively expressed by tumours seemingly insensitive to these cell cycle arrest genes given their Ki67 + nature (Fig. 3i and Extended Data Figs. 6a and 7b,c ). Aqp5 , an aquaporin inductor of gastric and colon carcinogenesis 26 , marked specific tumours able to produce the oncogenesis-promoting fibroblast growth factor FGF13 (Extended Data Fig. 7b,c ). Together with other markers (Extended Data Fig. 7b ) and corroborations at the protein level (Extended Data Fig. 7d ), these data indicate that a variety of tumour subtypes can be generated in the mini-colon, arguably due to tumour-niche-intrinsic and/or environmental factors. This probably accounts for the observed differences among mini-colon AKP cell lines (Fig. 2g and Extended Data Fig. 4h ). Importantly, we found that this diversity was relatable to the human context. For example, mini-colons generated tumours with transcriptional profiles representing both iCMS2- and iCMS3-like subtypes 27 (Extended Data Fig. 8a,b ) that were associated with a wide range of aggressiveness profiles (Extended Data Fig. 8c ) and correlated with different extents of lymph node colonization (Extended Data Fig. 8d,e ) when cross-compared with transcriptomic data from the TCGA collection of patients with CRC. Collectively, these findings demonstrate that the mini-colon is a complex cellular ecosystem that recreates both healthy and cancer cell diversity.
Screening of tumorigenic factors
The longevity, experimental flexibility and tumour formation dynamics of mini-colons provides an unparalleled in vitro set-up for conducting tumorigenesis assays. We therefore next used mini-colons as screening tools for identifying molecules with a prominent role in tumour development. As our single-cell RNA-seq (scRNA-seq) analyses revealed Gpx2 overexpression in cancer stem cells (Fig. 3g,h ), we probed its functional relevance by adding the glutathione peroxidase inhibitor tiopronin 28 to the basal medium reservoirs of mini-colons right after blue-light-induced AKP mutagenesis (Fig. 4a ). Basal application of the drug provides ubiquitous exposure on the mini-colon basolateral domain, mimicking a systemic therapy model (Fig. 4a ). By the time control mini-colons developed full-blown tumours, tiopronin-treated counterparts were largely tumour-free with a healthy colonic epithelium (Fig. 4b and Extended Data Fig. 9a ). This was not due to the mere reduction in proliferative activity, as tiopronin had a minor impact on organoid growth (Extended Data Fig. 9b,c ). As tiopronin targets several glutathione peroxidases, we corroborated the specific implication of GPX2 in tumour initiation by knocking down its transcript (Extended Data Fig. 9d ). These knockdown cells showed no detectable defects in terms of organoid morphology or proliferation in unchallenged conditions (Extended Data Fig. 9e,f ). However, after blue-light-mediated oncogenic recombination, GPX2-deficient mini-colons developed tumours with reduced kinetics and multiplicity (Fig. 4c and Extended Data Fig. 9g ), recapitulating the results obtained with tiopronin (Fig. 4b and Extended Data Fig. 9a ). Importantly, mini-colons were instrumental for these findings, as conventional organoid cultures cannot reveal differences in tumour-forming abilities (Extended Data Fig. 9b,h ).
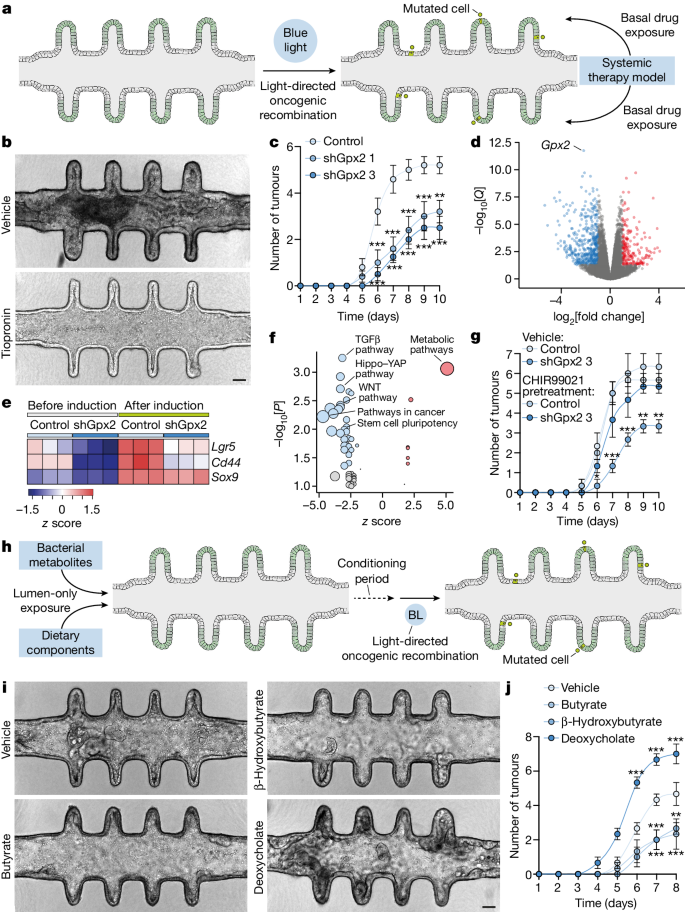
a , The experimental workflow for systemic therapy modelling. b , Bright-field images of mini-colons treated with vehicle or tiopronin after tumorigenic recombination. Images correspond to 6 days after induction. Scale bar, 75 μm. c , The multiplicity of tumours emerged in mini-colons of the indicated genotype after oncogenic induction. Statistical analysis was performed using two-way ANOVA with Sidak’s multiple-comparison test; ** P = 0.0034, *** P = 0.0007 (days 6 and 9, sh Gpx2 1), *** P < 0.0001 (all other conditions). n = 5, 5 and 4 mini-colons for control, shGpx2 1 and shGpx2 3, respectively. d , Differentially expressed genes after Gpx2 knockdown in light-induced AKP tumour cells. e , Expression of the indicated genes in colonocytes of the indicated genotypes before and after oncogenic recombination. The colour scale shows the z score. f , The main enriched functional terms after Gpx2 knockdown in light-induced AKP tumour cells. Significant terms are highlighted in blue or red, as determined using one-sided Fisher’s exact tests, with gene expression adjusted P -values (Benjamini-Hochberg correction). g , The multiplicity of tumours emerged in mini-colons of the indicated genotype under the indicated pretreatment (2 days before oncogenic induction). Statistical analysis was performed using two-way ANOVA with Sidak’s multiple-comparison test; * P = 0.0274, ** P = 0.0033 (days 9 and 10), *** P < 0.0001 (days 7 and 8). n = 3 mini-colons for each condition. h , The experimental workflow for microbiota and dietary pattern modelling. BL, blue light. i , Bright-field images of mini-colons treated with the indicated metabolites. Images correspond to 7 days after tumorigenic induction. Scale bar, 75 μm. j , The multiplicity of tumours emerged in mini-colons treated with the indicated metabolites. Statistical analysis was performed using two-way ANOVA with Sidak’s multiple-comparison test; ** P = 0.0080, *** P = 0.0008 (days 7 and 8), *** P < 0.0001 (day 6). n = 3 mini-colons for each condition. For c , g and j , data are mean ± s.e.m.
To gain molecular insights into the mechanism engaged by GPX2, we performed RNA-seq analysis of Gpx2 -knockdown cells both before and after oncogenic recombination. These analyses revealed that GPX2 deficiency remodels the colonocyte transcriptome in both healthy (Extended Data Fig. 9i ) and tumorigenic (Fig. 4d ) conditions (Supplementary Tables 1 and 2 ). This included the downmodulation of canonical markers associated with both healthy and cancer cell stemness, such as Lgr5 and Cd44 (Fig. 4e ). By contrast, markers of proliferative progenitor cells, such as Sox9 , remained unchanged (Fig. 4e ). Consistent with this, Gpx2 abrogation led to the repression of transcriptional programs implicated in stem cell pluripotency, including the WNT, Hippo–YAP and TGFβ pathways, as well as epithelial–mesenchymal transition and other processes involved in cancer cell fitness (Fig. 4f and Extended Data Fig. 9j,k ). Conversely, transcriptional programs associated with proliferation were not affected, consistent with our observations in cell culture (Extended Data Fig. 9e,f,k ). These findings indicate that the inhibition of GPX2 downmodulates colonocyte stemness, which probably accounts for the reduced tumorigenic potential observed in the mini-colon after oncogenic recombination. Supporting this, we found that non-transformed GPX2-deficient cells displayed reduced clonogenic capacity in medium deprived of exogenous WNT signals (Extended Data Fig. 9l,m ). Furthermore, the enhancement of WNT signalling through pretreatment of mini-colons with CHIR99021 for 2 days before oncogenic induction rescued the tumorigenic potential of Gpx2- knockdown cells (Fig. 4g and Extended Data Fig. 9n ). Collectively, these data uncover GPX2 as a key regulator of colon stemness and tumorigenesis, shedding light on lingering questions spurred by the recent discovery of its association with the malignant progression of human CRC 24 .
Besides cell-intrinsic factors, colon tumorigenesis in vivo is heavily modulated by a myriad of environmental molecules that continuously contact the luminal side of colonocytes, such as the metabolites produced by colon-residing microbiota 29 . The impact of these molecules cannot be faithfully evaluated in conventional organoid cultures, as their lumen is not accessible. As mini-colons address this limitation, we also investigated whether they could model the role of bacterial metabolites of which the tumorigenic function has been corroborated in vivo. To that end, we administered specific metabolites exclusively in the luminal side of healthy mini-colons and, after a conditioning period of 2 days, induced oncogenic recombination (Fig. 4h ). When luminally exposed to deoxycholic acid, a tumour-promoting metabolite 29 , 30 , 31 , mini-colons developed tumours with fast kinetics and high multiplicity (Fig. 4i,j ). Conversely, both tumour-suppressive butyrate 29 , 32 and β-hydroxybutyrate 33 slowed tumour development and reduced multiplicity (Fig. 4i,j ). These results demonstrate that mini-colons faithfully recapitulate the in vivo pathophysiological responses to bacterial metabolites, whereas conventional organoid cultures do not provide informative data on their tumorigenic relevance (Extended Data Fig. 10a ).
Dietary components also constitute a relevant source of luminal factors conditioning colon tumorigenesis 34 . We therefore performed analogous experiments modelling diets with different caloric contents (Fig. 4h and Extended Data Fig. 10b ). These revealed that calorie restriction in the luminal space effectively reduced tumour burden when compared to calorie-enriched medium (Extended Data Fig. 10c,d ), consistent with in vivo evidence 35 . To show the relevance of luminal accessibility, we placed the same amount of dietary medium in the basal medium reservoirs instead of the luminal space (Extended Data Fig. 10b ). Here, no differences were observed between the two dietary patterns (Extended Data Fig. 10e,f ), therefore indicating that an accessible lumen—a forbidden feature in conventional organoids—is decisive for the physiologically relevant modelling of colon biology. Collectively, these findings demonstrate that the mini-colon is a versatile tool that enables faithful in vitro recapitulation of CRC tumorigenesis and its environmental determinants.
Here we show that the mini-colon model shifts the paradigm of cancer initiation research, allowing ex vivo tumorigenesis with unparalleled pathophysiological intricacy. Coupled with spatiotemporal control of oncogenesis, real-time single-cell resolution and broad experimental flexibility, this system opens new perspectives for in vitro screening of cellular and molecular determinants of cancer development. Supporting this, mini-colons faithfully reflect in vivo-like responses to microbiota-derived metabolites and dietary patterns. Likewise, our model can help in the discovery and validation of genetic targets and tumour-suppressive drugs, as illustrated by the finding that glutathione peroxidase inhibition abrogates CRC tumour development. This constitutes a major advance over conventional 3D culture systems like organoids and Transwell models, which can recapitulate isolated aspects of colon biology such as histopathological features 36 or apical accessibility 37 , respectively, but lack the all-round topobiological complexity required to allow tumour formation ex vivo. Although such complexity demands bioengineering expertise to generate mini-colons, we have provided a detailed protocol that makes this system widely adoptable across laboratories that are already familiar with conventional organoid cultures (Protocol Exchange 38 ; see Methods ).
As for most genetic models of CRC, our system is based on the simultaneous acquisition of several mutations, which does not fully recapitulate the sequential tumorigenic process that occurs in vivo 39 . We therefore acknowledge that adopting a stepwise mutational system will enhance the relevance of the mini-colon as a cancer initiation model. We are also aware that spatial transcriptomics approaches will improve our understanding of tumour heterogeneity in the mini-colon. In the same lines, we envision the incorporation of additional regulatory layers in our OptoCre system, such as the fusion with the oestrogen receptor ligand-binding domain for subcellular localization control 40 , as a promising way to achieve finer spatiotemporal regulation of recombination.
Although mini-colons cannot be considered to be a general replacement for animals in all contexts of cancer research, they offer the possibility to reduce animal use in a wide range of experimental applications. Importantly, the pathophysiological relevance of the mini-colon can be readily enhanced by including stromal cells in the surrounding biomimetic extracellular matrix, which condition both tumour dynamics and invasiveness (Extended Data Fig. 10g–k ). Current lines of work that will be made available in ensuing publications have also proved that this model can be applied to patient-derived colorectal cancer specimens. Lastly, we anticipate that, by adapting its biomechanical properties, topology and culture conditions, it will be possible to expand the system to other prominent epithelial cancer types, such as lung, breast or prostate, bringing an important experimental resource to multiple fields.
Apc fl/fl mice (a gift from T. Petrova) were crossed to Cdx2-cre ERT2 mice (The Jackson Laboratory). Apc fl/fl Cdx2-cre ERT2 mice (termed A) were then crossed with Kras LSL-G12D/+ Trp53 fl/fl mice (a gift from E. Meylan) to generate Apc fl/fl Kras LSL-G12D/+ Trp53 fl/fl Cdx2-cre ERT2 mice (termed AKP). AKP mice were then back-crossed with C57BL6/J (The Jackson Laboratory) to generate Apc fl/fl Kras LSL-G12D/+ Cdx2-cre ERT2 mice (termed AK).
To induce tumorigenesis in vivo, Cre ERT2 recombinase was activated at the age of 8–10 weeks by a single intraperitoneal injection of 18 mg kg –1 tamoxifen (Sigma-Aldrich, T5648) in sunflower oil. Tumours were allowed to develop for 6 weeks. Mice were then sacrificed for tissue and cell isolation. See also below the specific section for transplantation of organoids in immunocompromised mice.
All animal work was conducted in accordance with Swiss national guidelines, reviewed and approved by the Service Veterinaire Cantonal of Etat de Vaud (VD3035.1 and VD3823). These regulations established 800 mm 3 as the maximal subcutaneous tumour volume allowed, which was not exceeded in any of the experiments. In experiments in which tumorigenesis was induced in vivo, the locomotion, appearance, body condition and intestinal function of the mice were monitored twice weekly and assigned numerical scores to allow quantitative decision making in case humane end points were necessary before the predefined end point of the experiment (6 weeks). All of the mice in this study reached the predefined end point. Mice were kept in the animal facility under EPFL animal care regulations. They were housed in individual cages at 23 ± 1 °C and 55 ± 10% humidity under a 12 h–12 h light–dark cycle. All of the animals were supplied with food and water ad libitum.
OptoCre module plasmid generation
The OptoCre module was designed by integrating the following constructs: (1) FUW-M2rtTA, which constitutively expresses the reverse tetracycline transactivator (rtTA); (2) FUW-tetO-GAVPO, which expresses the light-switchable trans-activator GAVPO after rtTA binding in the presence of doxycycline; and (3) FUW-OptoCre, which expresses Cre recombinase after GAVPO binding in the presence of blue light (Extended Data Fig. 1a ). FUW-M2rtTA was purchased from Addgene (20342). Vectors containing GAVPO and the GAVPO-binding promoter (UASG) 5 -P min , developed previously 41 , were a gift from M. Thomson 42 . For FUW-tetO-GAVPO generation, GAVPO was subcloned into the doxycycline-responsive FUW-TetO backbone (Wernig Lab, Stanford) using the EcoRI and NheI restriction sites (Extended Data Fig. 1a ). For FUW-OptoCre generation, (UASG) 5 -P min was inserted into the FUW-TetO backbone from which the TetO promoter had been removed (Wernig Lab, Stanford) using the BstBI and BamHI restriction sites. We then introduced the Cre recombinase (Addgene, 25997) downstream of (UASG) 5 -P min using the Pac1 restriction sites (Extended Data Fig. 1a ).
Isolation of colon cells
Healthy colon or tumour pieces were finely chopped using a scalpel and transferred to a gentle-MACS C-tube (Miltenyi, 130-093-237) containing 4 ml of digestion medium (RPMI (Thermo Fisher Scientific, 22400089), 1 mg ml –1 collagenase type IV (Life Technologies, 9001-12-1), 0.5 mg ml –1 dispase II (Life Technologies, 17105041) and 10 μg ml –1 DNase I (Applichem, A3778)). Tissues were then digested using the 37C_m_TDK_1 program on the gentle-MACS Octo Dissociator with heaters (Miltenyi). After the program was complete, the cell suspension was passed through a 70-μm strainer (Corning, 431751) and centrifugated at 400 g for 5 min.

Organoid and stromal cell culture
To establish organoids, colon cells were embedded in growth-factor-reduced Matrigel (Corning, 356231) (~2 × 10 4 cells per 20 μl dome) and cultured in Advanced DMEM/F-12 (Thermo Fisher Scientific, 12634028) supplemented with 1× GlutaMax (Thermo Fisher Scientific, 35050038), 10 mM HEPES (Thermo Fisher Scientific, 15630056), 100 μg ml −1 penicillin–streptomycin (Thermo Fisher Scientific, 15140122), 1× B-27 supplement (Thermo Fisher Scientific, 17504001), 1× N2 supplement (Thermo Fisher Scientific, 17502001), 1 mM N -acetylcysteine (Sigma-Aldrich, A9165), 50 μg ml −1 primocin (InvivoGen, ant-pm-2), 50 ng ml −1 EGF (Peprotech, 315-09), 100 ng ml −1 noggin (produced at EPFL Protein Production and Structure Core Facility), 500 ng ml −1 R-spondin (produced at EPFL Protein Production and Structure Core Facility), 50 ng ml –1 WNT3A (Time Bioscience, rmW3aL-010), 10 mM nicotinamide (Calbiochem, 481907) and 2.5 μM Thiazovivin (Stemgen, AMS.04-0017). This full medium is termed ‘WENRNi’. The base version of this medium without EGF, noggin, R-spondin, WNT3A and nicotinamide is referred to as BMGF and was used for the expansion of colon tumour organoids since they do not require the additional growth factors. The base version of BMGF without B-27, N2 and N -acetylcysteine is termed BM or basal medium, and was used for growth-factor deprivation experiments. A detailed protocol describing organoid culture can be found elsewhere 9 . Where indicated, organoids were treated with the following compounds or growth factors: regorafenib (8 μM, Selleckchem, S1178), ripretinib (1 μM, Selleckchem, S8757), infigratinib (1 μM, Selleckchem, S2183), SCF (100 ng ml –1 , PeproTech, 250-03), FGF2 (50 ng ml –1 , Thermo Fisher Scientific, PMG0035) and IGF1 (100 ng ml –1 , R&D Systems, 291-G1-200). Stromal cells were derived from cell suspensions from the primary tissue cultured in EGM-2 MV Microvascular Endothelial Cell Growth Medium-2 (Lonza, CC-3202) on conventional cell culture flasks. This medium selection strategy was followed by magnetic-activated cell sorting (MACS) on EPCAM (Miltenyi Biotec, 130-105-958) according to the manufacturer’s instructions to discard epithelial cells. The presence of stromal cells was further confirmed by immunofluorescence analyses of vimentin expression (see below). Cells were tested for mycoplasma before cryopreservation and in randomized routine checks using the MycoAlert PLUS Mycoplasma Detection Kit (Lonza, LT07-705).
Generation of light-inducible cells
Lentiviral particles carrying the three components of the OptoCre module (see above; Extended Data Fig. 1b ) and a Cre recombination reporter were produced at the EPFL Gene Therapy Platform by transfecting HEK293 cells with each plasmid of the OptoCre module and pLV-CMV-LoxP-DsRed-LoxP-eGFP (Addgene, 65726) plasmids. Lentivirus-containing supernatants were collected and concentrated by centrifugation (1,500 g for 1 h at 4 °C). Lentiviral titration was performed using the p24-antigen ELISA (ZeptoMetrix, 0801111). For transduction, colon organoids (around 2 × 10 5 cells) were dissociated into single cells by incubating in TrypLE Express Enzyme (Thermo Fisher Scientific, 12605028) at 37 °C for 5 min. Cells were then washed with basal medium supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, 10500064) and resuspended in WENRNi medium containing 8 μg ml −1 polybrene (Sigma-Aldrich, TR-1003-G) and the following amounts of viral particles: ~10 ng of p24 FUW-M2rtTA per ml, ~80 ng of p24 FUW-tetO-GAVPO per ml, ~80 ng of p24 FUW-OptoCre per ml and ~1,000 ng of p24 CMV-LoxP-DsRed-LoxP-eGFP per ml. These cells were plated in a 24-well plate, centrifuged at 600 g for 60 min at room temperature, and incubated for 6 h at 37 °C. After incubation, the cells were collected, centrifuged, plated in 20 μl Matrigel domes in a 24-well plate and cultured in WENRNi medium. Cells expressing the Cre recombination reporter were selected by supplementing WENRNi medium with 8 μg ml −1 puromycin (InvivoGen, ant-pr-1).
Light-mediated oncogenic recombination
The OptoCre module requires (1) doxycycline to induce rtTA-mediated GAVPO expression and (2) blue light to induce GAVPO-mediated Cre recombinase expression (Extended Data Fig. 1a,b ). At the desired time of oncogenic induction, 2 μg ml −1 doxycycline hydrochloride (Sigma-Aldrich, D3072) was added to the culture medium of either the organoids or mini-colons. Light induction was then performed using a custom-made LightBox built by Baur SA and the Instant Lab at EPFL. The LightBox consisted of an Acqua A5 System (Acme Systems) that could be remotely parametrized using a custom-made web-based application. Communication between the Acqua A5 System and the microcontroller (PJRC, Teensy 3.2) was done through Blocky programming, which allowed for control of the LED drivers (Sparkfun, PicoDuck). The LEDs (Cree LEDs, XLamp XP-C Blue LEDs) were placed into a custom multilayer 24-well plate holder made of black anodized aluminium and polyphenylsulfone; the height was optimized for homogeneous light distribution within each well. The entire LightBox, plate-holder, LEDs and cables were made to be placed in the incubator (watertight and heat resistant). Diffusive elements (Luminit, Light Shaping Diffuser 80°) were used to render the illumination more homogeneous inside each well. The intensity of the blue light (450–465 nm, peak at 455 nm) was optimized, set to 100 μW cm −2 and shined on the cells for 3 h. After blue-light exposure, doxycycline was removed by washing the cultures with fresh medium. In experiments targeting the light to specific regions of the mini-colon, work was carried out in the dark using a near infrared light (Therabulb, NIR-A) to prevent leaky Cre expression. Light-targeting was performed using a photomask that was adapted to the dimensions of the mini-colon and that was created from a photoresist and chrome-coated standard 5 × 5 inch silica plate (Nanofilm) with an automated machine (VPG200 Heidelberg Instrument, 2.0 µm resolution). Once the exposed photoresist was developed, the chrome layer was wet-etched and the remaining photoresist was stripped using a mask processor (Hamatech HMR900) 9 .
Microdevice design, fabrication and loading
The microfluidic device used for mini-colon cultures was designed using Clewin 3.1 (Phoenix Software) and fabricated as previously described 9 . It was composed of three main compartments: (1) a hydrogel chamber for cell growth in the centre; (2) two basal medium reservoirs flanking the hydrogel compartment; and (3) inlet and outlet channels for luminal perfusion 9 . An extracellular matrix containing 80% (v/v) type I collagen (5 mg ml −1 , Reprocell, KKN-IAC-50) and 20% (v/v) growth–factor-reduced Matrigel was loaded into the hydrogel compartment. The microchannels constituting the mini-colon architecture within the hydrogel were designed using Adobe Illustrator CC 2019 and Wolfram Mathematica 11.3. They were then read by PALM RoboSoftware 4.6 (Zeiss) and ablated using a nanosecond laser system (1 ns pulses, 100 Hz frequency, 355 nm; PALM Micro-Beam laser microdissection system, Zeiss). The dimensions of the mini-colon architecture were described previously 9 . A detailed description of all the key steps required for the generation and maintenance of mini-guts is available at Protocol Exchange ( https://doi.org/10.21203/rs.3.pex-903/v1 ) 38 .
Mini-colon culture, development and tumorigenesis
Colon organoids were dissociated into single cells by incubating in TrypLE Express Enzyme for 5 min at 37 °C followed by vigorous pipetting. This cell suspension was washed in 5 volumes of Advanced DMEM/F-12 supplemented with 10% FBS and passed through 40 μm cell strainers (Corning, 431750). After centrifugation at 400 g for 5 min, cells were resuspended in WENRNi medium at around 10 6 cells per ml. The mini-colon luminal microchannel was filled with 10 μl of this cell suspension. Cells were allowed to settle down in the mini-colon crypt-shaped cavities for 5 min, and the leftover unadhered cells were washed out from the microchannel by medium perfusion. The basal medium reservoirs were filled with 100 μl of WENRNi. Unless otherwise indicated, once the healthy colonic epithelium was fully formed (around 2 days after seeding), the medium in the luminal channel was switched to BM, while WENRNi was kept in the basal medium reservoirs. This gradient of growth factor from basal medium reservoirs to luminal space favours colonocyte differentiation across the crypt–lumen axis. For low-differentiation conditions of the differentiation experiments, WENRNi was kept in both the lumen and basal medium reservoirs. Conversely, high-differentiation mini-colons were cultured in WENRNi medium without WNT3A and nicotinamide (termed ENR). Unless otherwise stated, once the colonic epithelium was fully formed, oncogenic induction in the mini-colons was performed as stated above. Where indicated, tiopronin (5 mM, Selleckchem, S2062) or CHIR99021 (3 μM, StemCell Technologies, 100-1042) was added to the basal medium reservoirs after or before oncogenic induction, respectively. For co-culture experiments, ~500 stromal cells were seeded in each hydrogel before the laser-mediated ablation of the mini-colon pattern. The rest of the culture conditions and procedures remained unchanged. To avoid potential unspecific results derived from the small (but non-zero; Extended Data Fig. 1d,e ) leakiness of the optogenetic system, each replication across all studies was performed using independent OptoCre organoid lines freshly generated before each experiment. In all cases, the mini-colons were incubated at 37 °C in 5% CO 2 humidified air, with daily luminal perfusions and medium changes every other day.
Mini-colon whole-mount immunofluorescence staining
Mini-colons were rinsed with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde (Thermo Fisher Scientific, 15434389) overnight at 4 °C. After rinsing with PBS, the hydrogels were extracted from the PDMS scaffold using a scalpel, placed into a 48-well plate, permeabilized with 0.1% Tween-20 (Sigma-Aldrich, P9416) in PBS (10 min at 4 °C) and blocked in 2 mg ml −1 bovine serum albumin (Sigma-Aldrich, A3059) in PBS containing 0.1% Triton X-100 (Sigma-Aldrich, T8787) (blocking buffer) for at least 45 min at 4 °C. The samples were subsequently incubated overnight at 4 °C in blocking buffer with the corresponding following primary antibodies: CD44 (1:200; Abcam, ab157107), FABP1 (1:100; R&D Systems, AF1565), SOX9 (1:200; Abcam, ab185966), GPX2 (1:200; Bioss Antibodies, BS-13396R), IL-1α (1:200; R&D Systems, AF-400-SP), CDKN2A (1:100; Abcam, ab211542), E-cadherin (1:100; Abcam, ab11512) and vimentin (1:200; Abcam, ab92547). After three washes in blocking buffer for a total of 6 h at room temperature, the samples were incubated overnight at 4 °C in blocking buffer with the following corresponding secondary antibodies: Alexa Fluor 488 anti-goat (1:400, Thermo Fisher Scientific, A-11055), Alexa Fluor 488 anti-rat (1:400, Thermo Fisher Scientific, A-21208) and Alexa Fluor 647 anti-rabbit (1:400, Thermo Fisher Scientific, A-31573). After 3 washes in blocking buffer for a total of 6 h at room temperature, the samples were incubated with DAPI (1 μg ml −1 ; Tocris Bioscience, 5748) for 10 min at room temperature in blocking buffer. Before imaging, the hydrogels were mounted onto 35 mm glass bottom dishes (Ibidi, 81218-200) in Fluoromount-G (SouthernBiotech, 0100-01).
Mini-colon sectioning and histochemistry
Mini-colons were fixed and extracted from the PDMS scaffold as indicated above and were prepared for cryosectioning by incubating in 30% (w/v) sucrose (Sigma-Aldrich, S1888) in PBS until the sample sank. Subsequently, the samples were incubated for 12 h in a mixture of Cryomatrix (Epredia, 6769006) and 30% sucrose (mixing ratio 50/50) followed by a 12 h incubation in pure Cryomatrix. The samples were then embedded in a tissue mould, frozen on dry ice, and cut into 40-µm-thick sections at −20 °C using the CM3050S cryostat (Leica). Haematoxylin and eosin staining was performed at the EPFL Histology Core Facility using the Ventana Discovery Ultra automated slide preparation system (Roche).
Microscopy and image analysis
Bright-field and fluorescence imaging of living organoids and mini-colons was performed using the Nikon Eclipse Ti2 inverted microscope with ×4/0.13 NA, ×10/0.30 NA and ×40/0.3 NA air objectives and a DS-Qi2 camera (Nikon Corporation). Time lapses were taken in a Nikon Eclipse Ti inverted microscope system equipped with ×4/0.20 NA and ×10/0.30 NA air objectives and DS-Qi2 (Nikon Corporation) and Andor iXon Ultra 888 (Oxford Instruments) cameras. Both systems were controlled using the NIS-Elements AR software (Nikon Corporation). The extended depth of field (EDF) of bright-field images was calculated using a built-in NIS-Elements function. Fluorescence confocal imaging of fixed mini-colons was performed using the Leica SP8 STED 3X inverted microscope system equipped with ×10/0.30 NA air and ×25/0.95 NA water objectives, 405 nm diode and supercontinuum 470–670 nm lasers, and the system was controlled by the Leica LAS-X software (v.3.5.7, Leica microsystems). Histological sections were imaged using a Leica DM5500 upright microscope with ×10/0.30 NA and ×20/0.75 NA air objectives, a ×40/1.0 NA oil objective and a DMC 2900 Color camera, and the system was controlled by the Leica LAS-X software. Image processing was performed using standard contrast- and intensity-level adjustments in ImageJ (NIH). For oncogenic recombination analyses, the GFP-positive area was measured from 16-bit EDF images by subtracting the background, sharpening the images, and applying a signal threshold and a mask. The ratio between GFP-positive area and total organoid area was used for analyses. Recombined cells were segmented using StarDist with the default parameters ( https://github.com/stardist ) on the GFP channel of mini-colon images. Cell debris was discarded from segmentation analyses by setting an empirically established size threshold. For tumour quantification in the mini-colon, neoplastic structures with at least three times the thickness of the surrounding healthy epithelium were considered to be tumours. Videos of immunostainings were rendered using Imaris (Oxford Instruments).
Mini-colon shedding evaluation
The medium from the luminal compartments of the mini-colons, together with an additional luminal perfusion of 10 μl of basal medium, was collected every day for 4 days after the blue-light-induced oncogenic recombination. The protein content in these extracts was analysed using conventional Bradford assays (Bio-Rad, 5000006) and used as an indicator of cell shedding.
Mini-colon cell line derivation
Mini-colon-containing hydrogels were extracted from their microfluidic devices with a scalpel as indicated above and incubated with 0.1% (w/v) collagenase I (Thermo Fisher Scientific, 17100-017) at 37 °C for 10 min. Once the hydrogel was fully digested, the mini-colon was washed with PBS and digested with TrypLE Express Enzyme for 5 min at 37 °C. The resulting cell suspension was washed with Advanced DMEM/F-12 supplemented with 10% FBS, pelleted, embedded in Matrigel and cultured as indicated above for regular colon organoids.
Transplantation of organoids in immunocompromised mice
Organoid lines were established as indicated above from either in vivo colon tumours (reference AKP) or tumour-bearing mini-colons (mini-colon AKP). These organoids were dissociated into single cells using TrypLE Express Enzyme for 5 min at 37 °C, washed with Advanced DMEM/F-12 supplemented with 10% FBS, pelleted and embedded in Matrigel at 2.5 × 10 6 cells per ml. A total of 100 μl of this suspension was inoculated by subcutaneous injection into the right flank of NOD. Cd-Prkdz scid Il2rg tm1Wjl /Szj (NSG) mice (Jackson laboratories). Tumour growth was monitored using callipers twice per week until the end point at 18 days after inoculation. Length ( L ) and width ( W ) were measured and used to approximate the volume ( V ) of the tumour in mm 3 using the modified ellipsoid formula: V = ( L × W 2 )/2. After euthanasia, tumours were resected from the graft location and measured once more with callipers.
Graft sectioning and histochemistry
Tumour samples were fixed overnight in 4% paraformaldehyde at 4 °C, dehydrated in graded ethanol baths, cleared with xylene, embedded in paraffin and cut into 4-µm-thick sections using the HM 325 Rotary Microtome (Thermo Fisher Scientific). These sections were mounted onto Superfrost plus slides (Epredia, J1800AMNZ) and allowed to dry for 2 days at room temperature. Haematoxylin and eosin staining was performed at the EPFL Histology Core Facility using the Ventana Discovery Ultra automated slide preparation system (Roche).
Mutational screening in colon organoids
Genomic DNA was isolated from colon cells using the PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific, K182001) according to the manufacturer’s instructions. Recombination of the LSL (LoxP-Stop-LoxP) cassette controlling Kras G12D expression was confirmed by PCR using the protocol and oligos described by the Tyler Jacks laboratory ( https://jacks-lab.mit.edu/ , Kras G12D Conditional PCR). Apc and Trp53 recombinations were confirmed through exome sequencing performed at BGI Genomics at 100× coverage using DNBSEQ sequencing technology. DNA reads were mapped to the mouse GRCm39 genome assembly using BWA-MEM (v.0.7.17), filtered using samtools (v.1.9) and visualized using IGV (Integrative Genomics Viewer, Broad Institute, v.2.12.3).
Organoid proliferation assays
Single-cell suspensions of colon cells were generated as indicated above and embedded in 10 μl Matrigel domes at around 10 4 cells per dome in a 48-well plate. For each of the following 4 days, 220 μM resazurin (Sigma-Aldrich, R7017) was added to the culture medium and incubated for 4 h at 37 °C. Next, the resazurin-containing medium was collected and replaced with regular medium. Organoid proliferation was estimated by measuring the reduction of resazurin to fluorescent resorufin in the medium each day using the Tecan Infinite F500 microplate reader (Tecan) with 560 nm excitation and 590 nm emission filters. In the case of colony-formation assays, seeding was performed at around 10 3 cells per dome and the resulting colonies were counted after 3 days.
Organoid RNA extraction and bulk transcriptome profiling
Before RNA isolation, organoids were cultured for 3 days as indicated above and starved for 24 h in BM for the evaluation of growth-factor dependence. In the case of the Gpx2- knockdown experiments, 2 timepoints were analysed: 0 and 2 weeks after blue-light-induced activation (before and after oncogenic recombination, respectively). In all cases, cells were collected using TrypLE Express Enzyme as indicated above and lysed in RLT buffer (Qiagen, 74004), and the RNA was extracted using the QIAGEN RNeasy Micro Kit (Qiagen, 74004) according to the manufacturer’s instructions. Purified RNA was quality checked using a TapeStation 4200 (Agilent), and 500 ng was used for QuantSeq 3′ mRNA-seq library construction according to the manufacturer’s instructions (Lexogen, 015.96). Libraries were quality checked using a Fragment Analyzer (Agilent) and were sequenced in the NextSeq 500 (Illumina) system using NextSeq vm2.5 chemistry with Illumina protocol 15048776. Reads were aligned to the mouse genome (GRCm39) using star (v.2.7.0e) 43 . R (v.4.1.2) was used to perform the differential expression analyses. Count values were imported and processed using edgeR 44 . Expression values were normalized using the trimmed mean of M values (TMM) method 45 and low-expressed genes (<1 counts per million) were filtered out. Differentially expressed genes were identified using linear models (Limma-Voom) 46 and P values were adjusted for multiple comparisons using the Benjamini–Hochberg correction method 47 . Volcano plots and heat maps were generated using the EnhancedVolcano ( https://github.com/kevinblighe/EnhancedVolcano ) and heatmap3 ( https://github.com/slzhao/heatmap3 ) packages, respectively. The in vivo AKP signature was established from the differentially expressed genes between in vivo and organoid AKP lines with a log 2 -transformed fold change of at least |2|. To evaluate the enrichment of the in vivo AKP gene expression program across samples, the enrichment scores for both the upregulated and downregulated signatures were calculated using single-sample GSEA (ssGSEA) 48 . The difference between the two normalized enrichment scores yielded the fit score. ssGSEA was also used to analyse the enrichment of the MSigDB curated Hallmark gene set 49 in Gpx2 -knockdown organoids. Functional annotation was performed using DAVID 50 on the genes with a log 2 -transformed fold change of at least |1|. GOplot 51 was used for the integration of expression and functional annotation data. Known functional interactions among relevant genes were obtained through STRING 52 . Cytoscape 53 was used to perform network data integration and visualization.
Single-cell transcriptome profiling and lineage tracing
Lineage tracing was performed using the CellTag system 22 (V1 pooled barcode library, Addgene, 115643-LVC). In brief, we co-transduced inducible colon organoids with the CellTag barcode library (multiplicity of infection of around 5) and the OptoCre module as indicated above. These cells were then introduced and induced in the mini-colon system as indicated before. After 7 days in the system and when mini-colon tumours were clearly visible, we extracted the cells from mini-colons as indicated above. After pooling and filtering (40 μm) the cell suspensions from two mini-colons, the single-cell sequencing library was constructed using 10x Genomics Chromium 3′ reagents v3.1 according to the manufacturer’s instructions (10x Genomics, PN-1000269, PN-1000127, PN-1000215). Sequencing was performed using NovaSeq 6000 v1.5 reagents (Illumina protocol #1000000106351 v03) for around 100,000 reads per cell. The reads were aligned using Cell Ranger (v.6.1.2) 54 to the mouse genome (mm10) carrying artificial chromosomes for both GFP and CellTag UTR genes, as recommended by CellTag developers for facilitating barcode identification 55 . Raw count matrices were imported into R and analysed using Seurat (v.4.2.0) 56 . Dead cells were discarded on the basis of the number of detected genes (less than 3,000) and the percentage of mitochondrial genes (more than 20%), leading to 2,429 cells after filtering. The data were log-normalized and scaled, and dimensionality reduction was conducted using UMAP with 10 dimensions. Louvain clustering yielded 17 clusters that were merged and named on the basis of canonical cell type markers. Stem, cycling, progenitor, goblet and enteroendocrine cell scoring was based on published signatures in mini-intestines and in vivo 9 . Gene sets highlighting bottom, middle and top colonocytes were taken from enterocyte zonation studies 23 . Cancer stemness was scored based on the expression of Lgr5 , Cd44 and Sox9 . Intrinsic consensus molecular subtype (iCMS) signatures for colorectal cancer were obtained from published work 27 . Signature scoring was performed using burgertools ( https://github.com/nbroguiere/burgertools ). Visual representations of the data were generated using Seurat internal functions. For lineage-tracing analyses, CellTag detection, quantification and clone calling were performed as indicated by CellTag developers 55 , excluding cells expressing fewer than 2 or more than 30 CellTags. After filtering, 83 clonal populations were identified, from which only those with a minimum size of 5 cells were considered for further analyses. To identify clonal populations belonging to tumour cells, we looked for cells expressing transcripts carrying the genetically engineered Apc and Trp53 mutations, that is, deletions of exons 15 and 2–10, respectively (Extended Data Figs. 3g and 6b,c ). Note that this approach could not be performed for Kras , as the mutation is also present in the transcripts from WT cells (but not expressed). As scRNA-seq provides low coverage on exon junctions and therefore the presence of mutations can be assessed only in a small fraction of cells, we used both the cell-type composition and size distributions of bona fide mutationally confirmed tumour clonal populations to classify the rest of clones. Those falling within plus or minus 2 s.d. of the mean cell composition and size of bona fide tumours were classified as tumour clonal populations. Healthy clones were defined as those with a clearly distinct (outside the aforementioned range) cell type composition and the same upper limit size as was observed for tumour clones. After filtering and classification, 16 healthy and 18 tumour clonal populations were obtained and used for further analyses (Extended Data Fig. 6d ). To define the most robust tumour-clone-specific markers, the gene expression from cells in each clone was compared to that from cells in each other clone using the Wilcoxon rank-sum test. We considered only the positive markers and selected those with adjusted P < 10 −5 . The association of these markers with clinical parameters in patients with CRC (survival, lymph node staging) was performed through cBioPortal ( https://www.cbioportal.org/ ) using the 640-sample CRC TCGA dataset ( https://www.cancer.gov/tcga ) and a differential expression threshold equal or greater than |2 |. Further information is provided in the Data availability and Code availability sections.
shRNA-mediated transcript knockdown
Organoids were transduced as indicated above with lentiviral particles encoding Gpx2 shRNAs obtained from Sigma-Aldrich (TRCN0000076529, TRCN0000076531 and TRCN0000076532; sh Gpx2 1, sh Gpx2 2 and sh Gpx2 3, respectively) or, as a control, shRNA-free counterparts (Addgene, 65726). Transduced cells were selected with puromycin (5 μg ml −1 ; InvivoGen, ant-pr-1). Proper transcript knockdown was assessed using quantitative PCR with reverse transcription (RT–qPCR) and RNA-seq.
Analysis of mRNA abundance
Organoids were cultured and collected as indicated above. Cells were then lysed in RLT buffer and RNA was extracted using the QIAGEN RNeasy Micro Kit as indicated above. RT–qPCR was performed using the iTaq Universal SYBR Green One-Step Kit (Bio-Rad Laboratories, 1725150) and the QuantStudio 7 Flex Real-Time PCR System (Thermo Fisher Scientific, 4485701). Raw data were analysed using Design & Analysis Software (v.2.6.0, Thermo Fisher Scientific). We used the abundance of the endogenous Gapdh mRNA as internal normalization control. The following primers were used for transcript quantification: 5′-AGTTCGGACATCAGGAGAACTG-3′ (forward, Gpx2 ), 5′-GATGCTCGTTCTGCCCATTG-3′ (reverse, Gpx2 ), 5′-ATCCTGCACCACCAACTGCT-3′ (forward, Gapdh ) and 5′-GGGCCATCCACAGTCTTCTG-3′ (reverse, Gapdh ).
Microbiota and diet modelling
Inducible mini-colons were generated as indicated above. Once the epithelium was formed and before oncogenic induction, mini-colons were subjected to a conditioning period of 2 days in which luminal medium was (1) supplemented with 100 μM deoxycholate (Sigma-Aldrich, D2510), 10 mM butyrate (Sigma-Aldrich, B5887) or 10 mM β-hydroxybutyrate (Sigma-Aldrich, 54965); or (2) replaced with MEMα (calorie-restricted condition, Thermo Fisher Scientific, 22561-021) or Advanced DMEM/F12 supplemented with 30 μM palmitic acid (calorie-enriched condition, Sigma-Aldrich, P0500). The same concentrations were used in organoid control experiments, but these were added to the full culture medium as the luminal compartment is not accessible in organoids. To assess the relevance of luminal exposure to these factors in the mini-colon, the same total amounts were added in the basal medium reservoirs instead of the luminal channel. In all cases, after conditioning, oncogenic recombination was performed and the mini-colon was cultured as indicated above. The different medium compositions were replenished every day during luminal perfusion.
Statistics and reproducibility
The number of biological replicates ( n ), the type of statistical tests performed and the statistical significance for each experiment are indicated in the corresponding figure legend. For images associated with quantification charts (Fig. 1b,c with Fig. 1e ; Fig. 2b with Fig. 2c ; Fig. 2d with Extended Data Fig. 4b ; Fig. 2f with Fig. 2g ; Fig. 4b with Extended Data Fig. 9a ; Fig. 4i with Fig. 4j ; Extended Data Fig. 2a with Fig. 1e ; Extended Data Fig. 3d,e with Extended Data Fig. 3b ; Extended Data Fig. 3h with Fig. 1e ; Extended Data Fig. 5b with Extended Data Fig. 5c ; Extended Data Fig. 5d with Extended Data Fig. 5e ; Extended Data Fig. 9b with Extended Data Fig. 9c ; Extended Data Fig. 9e with Extended Data Fig. 9f ; Extended Data Fig. 9g with Fig. 4c ; Extended Data Fig. 9l with Extended Data Fig. 9m ; Extended Data Fig. 9n with Fig. 4g ; Extended Data Fig. 10c with Extended Data Fig. 10d ; Extended Data Fig. 10e with Extended Data Fig. 10f ), the number of replicates is the same as for the corresponding chart and is indicated in the figure legend of the latter. For the rest of representative images (Figs. 1d and 3h and Extended Data Figs. 1f , 2b,c,f , 3a , 4a , 7d , 9h and 10a,g–k ), three independent experiments were performed. scRNA-seq (Fig. 3a ) and exome sequencing with matched PCR (Extended Data Fig. 3f,g ) were performed with two independent sets of samples. Bulk RNA-seq was performed with at least three independent sets of samples. Unless otherwise indicated, statistical analyses were performed using GraphPad Prism v.9 (GraphPad). Data normality and equality of variances were analysed with Shapiro–Wilk and Bartlett’s tests, respectively. Parametric distributions were analysed using the Student’s t -test (when comparing two experimental groups) or ANOVA followed by either Dunnett’s test (when comparing more than two experimental groups with a single control group) or Tukey’s HSD test (when comparing more than two experimental groups with every other group). Nonparametric distributions were analysed using either Mann–Whitney U -tests (for comparisons of two experimental groups) or the Kruskal–Wallis followed by Dunn’s test (for comparisons of three or more than three experimental groups) tests. Sidak’s multiple-comparison test was used when comparing different sets of means. χ 2 tests were used to determine the significance of the differences between expected and observed frequencies. In all cases, values were considered to be significant when P ≤ 0.05. Data obtained are given as the mean ± s.e.m.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Bulk and single-cell RNA-seq data reported in this paper have been deposited at the Gene Expression Omnibus (GEO) public repository under accession number GSE221163 . The association analysis with clinical parameters in patients with CRC was performed through cBioPortal ( https://cbioportal.org ) using the 640-sample CRC TCGA dataset ( https://cancer.gov/tcga ). Source data are provided with this paper.
Code availability
The code used for data analysis is available at GitHub ( https://github.com/LorenzoLF/Mini-colon_bioengineering ) 57 and Zenodo ( https://doi.org/10.5281/zenodo.10057882 ) 58 .
Drost, J. & Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 18 , 407–418 (2018).
Article CAS PubMed Google Scholar
Rodrigues, J., Heinrich, M. A., Teixeira, L. M. & Prakash, J. 3D in vitro model (r)evolution: unveiling tumor-stroma interactions. Trends Cancer 7 , 249–264 (2021).
Tuveson, D. & Clevers, H. Cancer modeling meets human organoid technology. Science 364 , 952–955 (2019).
Article ADS CAS PubMed Google Scholar
Katt, M. E., Placone, A. L., Wong, A. D., Xu, Z. S. & Searson, P. C. In vitro tumor models: advantages, disadvantages, variables, and selecting the right platform. Front. Bioeng. Biotechnol. 4 , 12 (2016).
Article PubMed PubMed Central Google Scholar
Lee-Six, H. et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature 574 , 532–537 (2019).
Vendramin, R., Litchfield, K. & Swanton, C. Cancer evolution: Darwin and beyond. EMBO J. 40 , e108389 (2021).
Article CAS PubMed PubMed Central Google Scholar
Kim, J., Koo, B. K. & Knoblich, J. A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21 , 571–584 (2020).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459 , 262–265 (2009).
Nikolaev, M. et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 585 , 574–578 (2020).
Article ADS PubMed Google Scholar
Krotenberg Garcia, A. et al. Active elimination of intestinal cells drives oncogenic growth in organoids. Cell Rep 36 , 109307 (2021).
Liu, X. et al. Tumor-on-a-chip: from bioinspired design to biomedical application. Microsyst. Nanoeng. 7 , 50 (2021).
Article ADS PubMed PubMed Central Google Scholar
Augustine, R. et al. 3D bioprinted cancer models: revolutionizing personalized cancer therapy. Transl. Oncol. 14 , 101015 (2021).
Hubrecht, R. C. & Carter, E. The 3Rs and humane experimental technique: implementing change. Animals https://doi.org/10.3390/ani9100754 (2019).
Keum, N. & Giovannucci, E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 16 , 713–732 (2019).
Article PubMed Google Scholar
Bürtin, F., Mullins, C. S. & Linnebacher, M. Mouse models of colorectal cancer: past, present and future perspectives. World J. Gastroenterol. 26 , 1394–1426 (2020).
Drost, J. et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521 , 43–47 (2015).
Gehart, H. & Clevers, H. Tales from the crypt: new insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 16 , 19–34 (2019).
Levy, E. et al. Localization, function and regulation of the two intestinal fatty acid-binding protein types. Histochem. Cell Biol. 132 , 351–367 (2009).
Du, L. et al. CD44 is of functional importance for colorectal cancer stem cells. Clin. Cancer Res. 14 , 6751–6760 (2008).
Fleming, M., Ravula, S., Tatishchev, S. F. & Wang, H. L. Colorectal carcinoma: pathologic aspects. J. Gastrointest. Oncol. 3 , 153–173 (2012).
PubMed PubMed Central Google Scholar
Fujii, M. et al. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell 23 , 787–793 (2018).
Biddy, B. A. et al. Single-cell mapping of lineage and identity in direct reprogramming. Nature 564 , 219–224 (2018).
Article ADS CAS PubMed PubMed Central Google Scholar
Moor, A. E. et al. Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus axis. Cell 175 , 1156–1167 (2018).
Becker, W. R. et al. Single-cell analyses define a continuum of cell state and composition changes in the malignant transformation of polyps to colorectal cancer. Nat. Genet. 54 , 985–995 (2022).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143 , 134–144 (2010).
Kang, S. K. et al. Role of human aquaporin 5 in colorectal carcinogenesis. Am. J. Pathol. 173 , 518–525 (2008).
Joanito, I. et al. Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat. Genet. 54 , 963–975 (2022).
Hall, M. D. et al. Inhibition of glutathione peroxidase mediates the collateral sensitivity of multidrug-resistant cells to tiopronin. J. Biol. Chem. 289 , 21473–21489 (2014).
Louis, P., Hold, G. L. & Flint, H. J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 12 , 661–672 (2014).
Bernstein, C. et al. Carcinogenicity of deoxycholate, a secondary bile acid. Arch. Toxicol. 85 , 863–871 (2011).
Fu, T. et al. FXR regulates intestinal cancer stem cell proliferation. Cell 176 , 1098–1112 (2019).
Wu, X. et al. Effects of the intestinal microbial metabolite butyrate on the development of colorectal cancer. J. Cancer 9 , 2510–2517 (2018).
Dmitrieva-Posocco, O. et al. β-Hydroxybutyrate suppresses colorectal cancer. Nature 605 , 160–165 (2022).
Veettil, S. K. et al. Role of diet in colorectal cancer incidence: umbrella review of meta-analyses of prospective observational studies. JAMA Netw. Open 4 , e2037341 (2021).
Castejón, M. et al. Energy restriction and colorectal cancer: a call for additional research. Nutrients https://doi.org/10.3390/nu12010114 (2020).
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161 , 933–945 (2015).
Blutt, S. E. et al. Use of human tissue stem cell-derived organoid cultures to model enterohepatic circulation. Am. J. Physiol. Gastrointest. Liver Physiol. 321 , G270–G279 (2021).
Nikolaev, N. et al. Bioengineering microfluidic organoids-on-a-chip. Protocol Exchange https://doi.org/10.21203/rs.3.pex-903/v1 (2024).
Cho, K. R. & Vogelstein, B. Genetic alterations in the adenoma–carcinoma sequence. Cancer 70 , 1727–1731 (1992).
Meador, K. et al. Achieving tight control of a photoactivatable Cre recombinase gene switch: new design strategies and functional characterization in mammalian cells and rodent. Nucleic Acids Res. 47 , e97 (2019).
Wang, X., Chen, X. & Yang, Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods 9 , 266–269 (2012).
Sokolik, C. et al. Transcription factor competition allows embryonic stem cells to distinguish authentic signals from noise. Cell Syst. 1 , 117–129 (2015).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 , 15–21 (2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 , 139–140 (2010).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11 , R25 (2010).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15 , R29 (2014).
Reiner, A., Yekutieli, D. & Benjamini, Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19 , 368–375 (2003).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102 , 15545–15550 (2005).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) Hallmark gene set collection. Cell Syst. 1 , 417–425 (2015).
Dennis, G. et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4 , P3 (2003).
Walter, W., Sánchez-Cabo, F. & Ricote, M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 31 , 2912–2914 (2015).
Szklarczyk, D. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43 , D447–D452 (2015).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 , 2498–2504 (2003).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8 , 14049 (2017).
Kong, W. et al. CellTagging: combinatorial indexing to simultaneously map lineage and identity at single-cell resolution. Nat. Protoc. 15 , 750–772 (2020).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184 , 3573–3587 (2021).
Lorenzo-Martín, L. F. et al. Code for ‘Spatiotemporally resolved colorectal oncogenesis in mini-colons ex vivo’. GitHub github.com/LorenzoLF/Mini-colon_bioengineering (2024).
Lorenzo-Martín, L. F. et al. Code for ‘Spatiotemporally resolved colorectal oncogenesis in mini-colons ex vivo’. Zenodo https://doi.org/10.5281/zenodo.10057882 (2024).
Download references
Acknowledgements
We thank M. Thomson for the original light-inducible plasmids; M. Wernig and G. Neumayer for the initial idea and work on the doxycycline- and light-inducible system; C. Baur for discussing, designing and building the custom illumination device; A. Chrisnandy for assistance on photomask fabrication; O. Mitrofanova, B. Elci and Y. Tinguely for assistance on microdevice fabrication; D. Dutta and S. Li for input on organoid work; and J. Prébandier for administrative assistance. We acknowledge support from the following EPFL core facilities: CMi, CPG, PTBTG, HCF, BIOP, FCCF, BSF and GECF. This work was funded by the Swiss Cancer League (KFS-5103-08-2020), the Personalized Health and Related Technologies (PHRT) Initiative from the ETH Board and the EPFL.
Open access funding provided by EPFL Lausanne.
Author information
These authors contributed equally: L. Francisco Lorenzo-Martín, Tania Hübscher
Authors and Affiliations
Laboratory of Stem Cell Bioengineering, Institute of Bioengineering, School of Life Sciences and School of Engineering, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland
L. Francisco Lorenzo-Martín, Tania Hübscher, Nicolas Broguiere, Jakob Langer, Lucie Tillard & Matthias P. Lutolf
Swiss Institute for Experimental Cancer Research (ISREC), School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland
Amber D. Bowler & Freddy Radtke
Swiss Cancer Center Leman (SCCL), Lausanne, Switzerland
Institute of Human Biology (IHB), Roche Pharma Research and Early Development, Roche Innovation Center Basel, Basel, Switzerland
Mikhail Nikolaev & Matthias P. Lutolf
You can also search for this author in PubMed Google Scholar
Contributions
L.F.L.-M. conceived the study, designed experiments, performed the experimental and bioinformatic work, analysed the data, performed artwork design and wrote the manuscript. T.H. generated the OptoCre module and blue-light-associated systems, designed experiments, performed experimental work and analysed data. A.D.B. performed mouse-related work and isolated primary cells. N.B. performed bioinformatic work and analysed data. J.L. produced the microfluidic devices, optimized hydrogel patterning and generated mini-colon histological sections. L.T. performed experimental work. M.N. designed and developed the first mini-gut system. F.R. helped to conceive the work. M.P.L. conceived the work, designed experiments and edited the manuscript.
Corresponding authors
Correspondence to L. Francisco Lorenzo-Martín or Matthias P. Lutolf .
Ethics declarations
Competing interests.
The EPFL has filed for patent protection (EP16199677.2, PCT/EP2017/079651, US20190367872A1) on the scaffold-guided organoid technology used here, and M.P.L. and M.N. are named as inventors on those patents. M.P.L. is shareholder in Doppl, which is commercializing those patents. The other authors declare no competing interests.
Peer review
Peer review information.
Nature thanks Bradley Lega and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended data fig. 1 generation of blue–light-inducible akp colon organoids..
a , Schematic of the plasmids comprising the OptoCre module. The promoter, gene, and restriction sites used to generate the plasmids are indicated. b , Schematic of the integration of the different genetic elements that allow spatiotemporal control of oncogenic recombination in colon organoids. c , Schematic of the experimental workflow used to test and optimize the OptoCre system. d , Brightfield and fluorescence images of OptoCre-carrying inducible colon organoids exposed to the indicated conditions. Red and green signals correspond to healthy and mutated cells, respectively. Images were taken 48 h after induction. Scale bar, 200 μm. e , Recombination efficiency in inducible colon organoid exposed to the conditions indicated in panel d. *** P = 0.0001 (Kruskal-Wallis and Dunn’s multiple comparisons test; n = 9 cultures for each condition). Each point represents one well of organoids. Data represent mean ± SEM. f , Brightfield and fluorescence images of inducible colon organoids exposed to control (top) and activation (bottom) conditions, dissociated into single cells and replated in the absence of growth factors (BMGF medium). Green signal corresponds to mutated cells. Images were taken 24 h after replating. Scale bar, 200 μm.
Extended Data Fig. 2 Oncogenic mutations induce neoplastic growth in mini-colons.
a , Time-course brightfield and fluorescence images of non-induced healthy colon cells grown as conventional organoids and mini-colons. Absence of fluorescence signal indicates absence of oncogenic recombination. Scale bars, 200 μm (organoids) and 75 μm (mini-colons). b , Immunofluorescence images showing the expression of Fabp1 (left, green), and Sox9 (right, magenta) in healthy mini-colons cultured for 7 days. Scale bar, 100 μm. c , Fluorescence image (left) showing the presence of mutated cells 36 h after the blue–light-mediated induction of a mini-colon. The segmentation of each mutated cell is shown (right). Scale bar, 120 μm. d , Evolution of the number of mutated cells in an inducible mini-colon after blue–light-mediated activation. Each dot represents a measurement every 15 min. The 2 nd order smoothing of the data is shown. e , Time-course quantitation of cell shedding (total protein content) into the lumen of OptoCre and control mini-colons after blue-light induced oncogenic recombination. * P = 0.0156; ** P = 0.0033; *** P = 0.0003 (24 h), <0.0001 (96 h) (two-way ANOVA and Sidak’s multiple comparisons test; n = 3 mini-colons for each condition). Each point represents one mini-colon. Data represent mean ± SEM. f , Low- (left) and high-magnification (right) immunofluorescence images showing the presence of CD44 (magenta) and nuclei (blue) in a tumour-bearing mini-colon. White and grey arrowheads indicate early and advanced tumorigenic events, respectively. Scale bars, 120 μm (left) and 50 μm (right).
Extended Data Fig. 3 Oncogenic mutations induce full blown tumours in mini-colons.
a , Hematoxylin and eosin staining of a mini-colon tumour section. Scale bar, 25 μm. b , Time-course growth of tumours produced by cells derived from mini-colon tumours upon subcutaneous transplantation in immunodeficient mice ( n = 5 mice). As a reference, bona fide cancer cells from primary colon tumours are included. c , Image of the tumours at the endpoint of the experiment shown in panel b. d , Hematoxylin and eosin stainings of sections from the indicated tumour types. Zoomed-in areas (right) are indicated with a dashed square (left). Scale bar, 100 μm. e , Hematoxylin and eosin stainings of sections from mini-colon AKP implant tumours showing the presence of invading cancer cells (left, black arrowheads) and areas of cellular atypia (right, white arrowhead). Scale bar, 200 μm. f , Electrophoretic separation of PCR-amplified KRAS LSL locus in the indicated samples. See Methods for more details on PCR design. g , Whole exome sequencing coverage in the indicated loci and cells. Missing exons in recombined cells are indicated. h , Brightfield images of mini-colons of the indicated genotypes 23 days after blue light exposure. Neoplastic and tumour structures are indicated with black and white arrowheads in A and AK mini-colons, respectively. By that time tumours have extended throughout the whole mini-colon tissue in the case of the AKP model, forming a dense mass of cancer cells. Scale bar, 75 μm.
Extended Data Fig. 4 Mini-colon tumours display in vivo -like functional and transcriptional features.
a , Brightfield and fluorescence images of a mini-colon where blue light exposure has been targeted to a specific area (dashed blue line). Red and green signals correspond to healthy and mutated cells, respectively. Images were taken 36 h after induction. Scale bar, 75 μm. b , Multiplicity of tumours emerged in mini-colons cultured in the indicated conditions. * P = 0.0122; ** P = 0.0035; *** P = 0.0002 (two-way ANOVA and Sidak’s multiple comparisons test; n = 4 mini-colons for each condition). c , Distribution of tumour morphologies in mini-colons cultured in the indicated conditions. ** P = 0.0024 (two-way ANOVA and Sidak’s multiple comparisons test; n = 4 mini-colons for each condition). d , Brightfield images of the indicated colon organoid lines cultured for 3 days in full organoid medium. Scale bar, 200 μm. e , Metabolic activity (measured using resazurin) of the indicated colon organoid lines cultured in BMGF medium for the indicated time. Numeric labelling (1-8) is used to facilitate cell line identification. *** P = 0.0002 (mini-colon AKP #iii), <0.0001 (all other conditions) (two-way ANOVA and Sidak’s multiple comparisons test; n = 3 cultures for each line). f , Volcano plot of the differentially expressed genes between “in vivo” and “organoid AKP” cell lines. g , Top enriched functional clusters in the differentially expressed genes identified in panel f. h , Enrichment of the “in vivo AKP” transcriptional signature identified in panel f across the different “mini-colon” and “organoid AKP” lines. * P = 0.0137; ** P = 0.0032 (#iii), 0.0075 (#iv); *** P = 0.0001 (Brown-Forsythe ANOVA and Dunnett’s T3 multiple comparisons test (two-sided), n = 6 and 3 cultures for “organoid AKP” and the rest of cell lines, respectively). Each dot represents one culture. i , Main enriched functional terms in the differentially expressed genes between “mini-colon AKP” lines # i and # v. Significant terms are highlighted in red (one-sided Fisher’s exact test, adjusted P values). In b , c , e , and h , data represent mean ± SEM. GPL, glycerophospholipid; EL, ether lipid; PG, proteoglycans; SC, stem cell.
Extended Data Fig. 5 Tumorigenesis in the mini-colon leads to enhanced RTK signalling promoting growth factor independence.
a , Gene interaction network of the overlapping genes that are upregulated in AKP lines with high-proliferation potential in BM (“in vivo AKP”, “mini-colon AKP” #v) when compared to low-growth counterparts (“organoid AKP”, “mini-colon AKP” #i). Network hubs are highlighted with circles. b , Brightfield images of “mini-colon AKP” #v organoids cultured for 3 days with the indicated media and compounds. Scale bar, 200 μm. c , Metabolic activity (measured using resazurin) of “mini-colon AKP” #v organoids cultured in the indicated conditions. * P = 0.0236; *** P < 0.0001 (two-way ANOVA and Sidak’s multiple comparisons test; n = 4 cultures for infigranib and 8 for the rest of conditions). d , Brightfield images of “organoid AKP” cells cultured for 3 days in BM with the indicated growth factors. Scale bar, 200 μm. e , Metabolic activity (measured using resazurin) of “organoid AKP” cells cultured in the indicated conditions. *** P < 0.0001 (two-way ANOVA and Sidak’s multiple comparisons test; n = 8 cultures for each condition). In c and e , each point represents one well of organoids and data represent mean ± SEM.
Extended Data Fig. 6 Mini-colons comprise a complex cellular ecosystem.
a , Expression distribution of cell–type-specific markers across mini-colon cells. Cell-type labels can be found in Fig. 3b . b , Examples of single-cell RNA reads capturing exon-exon junctions that reveal the expected oncogenic recombination in Apc . c , Examples of single-cell RNA reads capturing exon-exon junctions that reveal the expected oncogenic recombination in Trp53 . d , Unsupervised UMAP clustering of the main cell types found in each of the healthy and tumour clonal populations found within the mini-colon. Tumour clones carry the “CRC” label. UMAP structure corresponds to the one shown in Fig. 3b .
Extended Data Fig. 7 Mini-colons display intra- and inter-tumour diversity.
a , Expression distribution of proliferation ( Mki67 ), stemness ( Cd44 ), and differentiation ( Krt20 ) markers within a single clonal tumour population. b , Heatmap of the genes showing the strongest ( P < 10 −5 ) differential expression across mini-colon tumours. The tumour clonal population is indicated on top. c , Expression of the indicated genes in the indicated tumour clones. *** P = 2.74·10 −14 ( Slpi , clone #1), 4.06·10 −50 ( Prdm16 , clone #14), 1.05·10 −13 ( Aqp5 , clone #25) (two-sided Wilcoxon rank-sum test; n = 540 cells). Each point represents one cell. d , Immunofluorescence images showing the expression of Il1a (left, green), Cdkn2a (right, red), and the presence of nuclei (cyan) in tumour-bearing and control mini-colons. White and grey arrowheads indicate positive and negative tumours, respectively, in terms of marker expression. Scale bar, 100 μm.
Extended Data Fig. 8 The tumour heterogeneity in the mini-colon is relatable to the human context.
a , Expression distribution of intrinsic consensus molecular subtype (iCMS) signatures across tumour cells in the mini-colon. Cell-type labels can be found in Fig. 3d . Exp, expression. b , Fraction of cells within each tumour clone in the mini-colon classified in each iCMS group. c , Survival of CRC patients from the TCGA database according to the expression of the indicated tumour clone-specific markers. The logrank test P value is indicated ( n = 375 patients). d , Presence of cancer cells in lymph nodes from CRC patients from the TCGA database according to the expression of the indicated tumour clone-specific markers. *** P = 2.714·10 −5 (two-sided Wilcoxon test; n = 375 patients). e , Lymph node staging in CRC patients from the TCGA database according to the expression of the indicated tumour clone-specific markers. * P = 0.0222; ** P = 4.923·10 −3 (two-sided Chi-squared test; n = 375 patients).
Extended Data Fig. 9 Gpx2 regulates colonocyte stemness and tumorigenesis.
a , Multiplicity of tumours emerged in mini-colons treated with the indicated compound upon oncogenic induction. *** P = 0.0001 (day 5), <0.0001 (all other conditions) (two-way ANOVA and Sidak’s multiple comparisons test; n = 6 mini-colons for each condition). b , Brightfield images of colon organoids treated with the indicated compound after tumorigenic recombination. Images correspond to 3 days after induction. Scale bar, 200 μm. c , Metabolic activity (measured using resazurin) of organoids cultured in the indicated conditions and times after oncogenic recombination. No significant differences (two-way ANOVA and Sidak’s multiple comparisons test; n = 4 cultures for each condition). d , qRT-PCR based quantitation of Gpx2 mRNA in the indicated cell lines. *** P < 0.0001 (one-way ANOVA and Tukey’s multiple comparisons test; n = 6, 4, and 3 organoid cultures for parental, sh Gpx2 #1, and the rest of the lines, respectively). e , Brightfield images of non-induced colon organoids of the indicated genotypes after 3 days of culture. Scale bar, 200 μm. f , Metabolic activity (measured using resazurin) of non-induced colon organoids of the indicated genotypes at the indicated times. No significant differences (two-way ANOVA and Sidak’s multiple comparisons test; n = 6 cultures for each condition). g , Brightfield images of mini-colons of the indicated genotypes after tumorigenic recombination. Images correspond to 6 days after induction. Scale bar, 75 μm. h , Brightfield images of colon organoids of the indicated genotypes after tumorigenic recombination. Images correspond to 6 days after induction. Scale bar, 200 μm. i , Volcano plot showing the differentially expressed genes upon Gpx2 knockdown in non-transformed colon cells. j , Bubble plot showing the main enriched functional terms in the differentially expressed genes upon Gpx2 knockdown in non-transformed colon cells. Significant terms are highlighted in either blue (downmodulated) or red (upmodulated) (one-sided Fisher’s exact test, adjusted P values). k , Enrichment of the indicated hallmark signatures from the MSigDB in the indicated cell lines. ** P = 0.0013; *** P = 0.0008 (Wnt), 0.0003 (EMT, before recombination), <0.0001 (all other conditions); NS, not significant (one-way ANOVA and Tukey’s multiple comparisons test; n = 3 cultures for each condition). l , Colony assay images of non-induced colon organoids of the indicated genotypes after 3 days of culture in the indicated media conditions. Scale bar, 200 μm. m , Clonogenic capacity of non-induced colon organoids of the indicated genotypes after 3 days of culture in the indicated media conditions. * P = 0.0219; ** P = 0.0012 (EN, sh Gpx2 #3), 0.0036 (BMGF, sh Gpx2 #1); *** P < 0.0001 (two-way ANOVA and Dunnet’s multiple comparisons test; n = 3 cultures for each condition). n , Brightfield images of Gpx2 knockdown mini-colons that had undergone the indicated pre-treatment before tumorigenic recombination. Images correspond to 7 days after tumour induction. Scale bar, 75 μm. In a , c , d , f , k , and m , each point represents one well of organoids and data represent mean ± SEM.
Extended Data Fig. 10 Mini-colons provide experimental versatility and resolution to tumorigenic studies.
a , Brightfield images of colon organoids treated with the indicated bacterial metabolites. Images correspond to 5 days after oncogenic induction. Scale bar, 200 μm. b , Schematic of the experimental setup used to evaluate the relevance of luminal access in tumorigenic studies. c , Brightfield images of mini-colons treated with the indicated diets according to experimental setup displayed in panel b (left). Images correspond to 6 days after tumorigenic induction. Scale bar, 75 μm. d , Multiplicity of tumours emerged in mini-colons treated with the indicated diets according to experimental setup displayed in panel b (left). *** P = 0.0001 (day 7), <0.0001 (days 6 and 8) (two-way ANOVA and Sidak’s multiple comparisons test; n = 4 and 3 mini-colons for calorie-restricted and -enriched diets, respectively). e , Brightfield images of mini-colons treated with the indicated diets according to experimental setup displayed in panel b (right). Images correspond to 6 days after tumorigenic induction. Scale bar, 75 μm. f , Multiplicity of tumours emerged in mini-colons treated with the indicated diets according to experimental setup displayed in panel b (right). Differences are not significant (two-way ANOVA and Sidak’s multiple comparisons test; n = 4 and 3 mini-colons for calorie-restricted and -enriched diets, respectively). g , Brightfield image of a healthy (non-transformed) mini-colon with integrated stromal cells in the extracellular matrix. Scale bar, 75 μm. h , Immunofluorescence image showing the presence of E-cadherin (green) and Vimentin (magenta) in the mini-colon shown in panel g. Scale bar, 75 μm. i , Brightfield images of mini-colons in the indicated mono- (left) and co-culture (right) setups. Images correspond to 6 days after tumorigenic induction. Scale bar, 75 μm. j , Brightfield image of an invasive front (arrowhead) formed in response to the presence of stromal cells in a mini-colon. The image corresponds to 6 days after tumorigenic induction (zoomed-in from panel i). Scale bar, 30 μm. k , Immunofluorescence image showing the presence of E-cadherin (green) and Vimentin (magenta) in the invasive front from panel j 23 days after tumorigenic induction. Scale bar, 20 μm. In d and f , data represent mean ± SEM.
Supplementary information
Reporting summary, supplementary table 1.
Differentially expressed genes in shGpx2 colon organoids before oncogenic recombination.
Supplementary Table 2
Differentially expressed genes in shGpx2 colon organoids after oncogenic recombination
Supplementary Video 1
Early response to oncogenic activation within a mini-colon. 46 h time-lapse video of mutated cells in a mini-colon 24 h after oncogenic recombination.
Supplementary Video 2
Hyperplasia and early tumour development in a mini-colon. 36 h time-lapse video of a mini-colon with multiple tumour-initiating events 5 days after oncogenic recombination.
Supplementary Video 3
Ex vivo tumour development in a mini-colon. 38 h time-lapse video of tumour development in a mini-colon 9 days after oncogenic recombination.
Supplementary Video 4
Cancer stem cells initiate tumour development in mini-colons. 3D visualization of cancer stem cell marker CD44 overexpression in early tumorigenic sites.
Supplementary Video 5
Intratumour complexity in mini-colons. 3D visualization of CD44 (cancer stem cell marker) and FABP1 (mature colonocyte marker) expression in mini-colon tumours and epithelium.
Source data
Source data fig. 1, source data fig. 2, source data fig. 3, source data fig. 4, source data extended data fig. 1, source data extended data fig. 2, source data extended data fig. 3, source data extended data fig. 4, source data extended data fig. 5, source data extended data fig. 8, source data extended data fig. 9, source data extended data fig. 10, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Lorenzo-Martín, L.F., Hübscher, T., Bowler, A.D. et al. Spatiotemporally resolved colorectal oncogenesis in mini-colons ex vivo. Nature (2024). https://doi.org/10.1038/s41586-024-07330-2
Download citation
Received : 02 February 2023
Accepted : 18 March 2024
Published : 24 April 2024
DOI : https://doi.org/10.1038/s41586-024-07330-2
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Mini-colon and brain 'organoids' shed light on cancer and other diseases.
- Sara Reardon
Nature (2024)
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Cancer newsletter — what matters in cancer research, free to your inbox weekly.

IMAGES
VIDEO
COMMENTS
Website Developmemt Technologies: A Review. Abstract: Service Science is that the basis of knowledge system and net services that judge to the provider/client model. This paper developments a technique which will be utilized in the event of net services like websites, net applications and eCommerce. The goal is to development a technique that ...
The paper gives an overview of the new features of web technologies. The general idea of the new version of HTML (Hyper Text Markup Language), i.e. HTML5, and other tools presented in this paper ...
This research paper discusses the process involved in developing a website in past and present, development of content delivery over the years, the website uses, a website for mobile devices, and performance comparison between two of the most used web backend development technologies, i. e, Node.js and Python.
Tegegne Tesfaye Haile. This research paper reviews the evolution of the Web from its early phase, web 1.0, to the present state, web 3.0. The primary emphasis is placed on the shift that users ...
The most basic and first step for development is to decide the right frontend framework. There is a wide variety of options emerging every year as solutions for the problems developers face every day with browser-based applications taking over the world. ... This paper initially discusses different trends in framework and libraries of ...
Abstract. This paper highlights current web development technologies in IT industry and its measure. Developers of web-based applications are confronted with a bewildering array of available choice formats, languages, frameworks, and technical objects. They investigate, identify, and evaluate technology for creating web applications.
Therefore, the present paper seeks to address the aforementioned deficiencies by providing an overview of the best practices on Web development including Lift's and implementing examples. 3. Best practices for Web development and comparison of Web frameworks in the use of best practices. A best practice is the best way to achieve an outcome.
Explore the latest full-text research PDFs, articles, conference papers, preprints and more on WEB APPLICATION DEVELOPMENT. Find methods information, sources, references or conduct a literature ...
The IS literature reveals considerable effort concerning the development of web-based systems. Particularly the differences and similarities between traditional systems development and web development and the applicability of traditional development methods are widely and controversially discussed. However, the discussions are still primarily ...
Web development is one of the ways to contribute to the digitization of organizations. However, being a relevant area, it has the contribution of countless companies and people worldwide. This high number of contributions necessarily leads to several trends and forms of development. This article explores one of these trends, namely the use of react on the frontend and nodejs on the backend. In ...
Rashid (2017) consider that there are five essential reasons that support the great growth of responsive design, respectively: (i) increased traffic from mobile users; (ii) lower cost and website maintenance; (iii) provides a seamless user experience; (iv) adapts easily to any screen size; and (v) improves SEO efforts.
Website development is becoming a growing concern for all organizations in this world. However the process and lifecycle involved in their development is still uncertain. The Web based system development usually involves more heterogeneous stakeholders as compared to traditional software's development construction. The process models used for the development of websites stands borrowed from ...
The research carried out by other authors and included in this study, serves as a fundamental approach for the comparison between the tools used by the development methodologies detailed in Table 9. Table 9. Comparison between web application development methodologies. Empty Cell.
Abstract. Pursuant to two broad "iterations" of the world wide web, the first relegating the user to a rudimentary read-only role, and the second to that of a content-creator bound by an architecture of oligopolistic entities, a third iteration (Web 3.0) is posited as one wherein the user is incarnated as the ultimate arbiter and primary agent of value creation and exchange.
2.3. Analysis. The literature review uncovered 20 distinct design elements commonly discussed in research that affect user engagement. They were (1) organization - is the website logically organized, (2) content utility - is the information provided useful or interesting, (3) navigation - is the website easy to navigate, (4) graphical representation - does the website utilize icons ...
accompany each server-side development and client-side development, full stack developers head the arrange of action and keep a track of the progress of the project. Within the finish of the paper, we will clearly outline Vision, Challenges and Future scope of Full Stack Web Development. Keywords: Full stack web development, Front-end,
Explore the latest full-text research PDFs, articles, conference papers, preprints and more on WEBSITE DEVELOPMENT. Find methods information, sources, references or conduct a literature review on ...
The responsive web design solves the compatibility problems of web pages displayed at different resolutions, different platforms, and different screen sizes, and also brings high-quality experience to users. Based on the research on responsive web design, and related technologies of HTML5 and CSS3, this paper expounds the design ideas and key technologies of responsive design with a responsive ...
An Evaluation Support Framework for Internet Technologies and Tools. In several stages of Web development, Web Engineers have to study, evaluate, compare and finally select one, among many, similar software systems/tools and theoretical Web resources, i.e. processes, methodologies, technologies and... more. Download. by Sotiris Christodoulou.
In particular, this paper is to review number of articles on web development and design related to diversity issues published according to year, to study databases contains 1 articles related to human diversity in web development and design, to identify theories that have been applied in the researches, to investigate method proposed by the ...
Oxfam is seeking a consultant(s) to assist in developing a White/Research Paper on AI Usage in Humanitarian and Development Sectors, with a particular emphasis on data empowerment and the creation ...
Full Stack Web Development: A Project-Based Learning Course Prepares Students for Industry 4.0 The abstract of a conference paper presented at The 7th Kinneret Conference on Software Engineering ...
Execute event-driven serverless code functions with an end-to-end development experience. Web App for Containers ... More details on benchmarks are provided in our technical paper. Note: Phi-3 models do not perform as well on factual knowledge benchmarks (such as TriviaQA) as the smaller model size results in less capacity to retain facts ...
Three-dimensional organoid culture technologies have revolutionized cancer research by allowing for more realistic and scalable reproductions of both tumour and microenvironmental structures1-3.
This paper starts with introducing an. overview of the leading frameworks and libraries in the field of. front-end development and examine each performance in web. services. By analyzing the ...
PDF | On Jan 25, 2021, Biplab Poudyal and others published Journal of Web Development and Web Designing | Find, read and cite all the research you need on ResearchGate.