
- Planet Earth
- Strange News

How To Write A Research Paper In Biotechnology
Table of Contents:
Current research in biotechnology: Exploring the biotech forefront . Biotechnology is an evolving research field that covers a broad range of topics. Here we aimed to evaluate the latest research literature, to identify…
Highlights – View PDFCurrent research in biotechnology: Exploring the biotech forefrontUnder a Creative Commons licenseopen accessHighlights•Biotechnology literature since 2017 was analyzed. •The United States of America, China, Germany, Brazil and India were most productive. •Metabolic engineering was among the most prevalent study themes. •Escherichia coli and Saccharomyces cerevisiae were frequently used. •Nanoparticles and nanotechnology are trending research themes in biotechnology. AbstractBiotechnology is an evolving research field that covers a broad range of topics. Here we aimed to evaluate the latest research literature, to identify prominent research themes, major contributors in terms of institutions, countries/regions, and journals. The Web of Science Core Collection online database was searched to retrieve biotechnology articles published since 2017. In total, 12,351 publications were identified and analyzed. Over 8500 institutions contributed to these biotechnology publications, with the top 5 most productive ones scattered over France, China, the United States of America, Spain, and Brazil.
Video advice: 7 Most Effective Tips For Writing a Better Scientific Research Paper
Thesis Writing, Research paper Writing, Research Methodology \u0026 Scientific Writing Course – https://btnk.org/btn-certification

BMC Biotechnology
BMC Biotechnology is an open access, peer-reviewed journal that considers articles on the manipulation of biological macromolecules or organisms for use in …
- Ethics approval and consent to participate
- Consent for publication
- Availability of data and materials
- Competing interests
- Authors’ contributions
- Acknowledgements
- Authors’ information
All manuscripts must contain the following sections under the heading ‘Declarations’: Ethics approval and consent to participate Consent for publication Availability of data and materials Competing interests Funding Authors’ contributions Acknowledgements Authors’ information (optional)Please see below for details on the information to be included in these sections. If any of the sections are not relevant to your manuscript, please include the heading and write ‘Not applicable’ for that section. Ethics approval and consent to participateManuscripts reporting studies involving human participants, human data or human tissue must: include a statement on ethics approval and consent (even where the need for approval was waived) include the name of the ethics committee that approved the study and the committee’s reference number if appropriateStudies involving animals must include a statement on ethics approval and for experimental studies involving client-owned animals, authors must also include a statement on informed consent from the client or owner.
Top Ten Exclusive Research Paper Topics On Biotechnology
Looking for some unique ideas for your paper on biotechnology? Check out the list of suggestions provided in the article and feel free to take your pick.
A Selection Of Great Research Paper Topics On Biotechnology – Like a student, you’ll frequently need to write complex academic assignments that need effort, search, critically planning and exploring new aspects. You are able to only produce a winning assignment if you opt to talk about fresh ideas and new breakthroughs. Its likely the first couple of topics which come for your mind under this subject could be already taken. You have to make certain the niche you decide to address is exclusive and fresh. If other scientific study has already spoken relating to this before you decide to, then there’s no reason on paper it.
Free Term Papers On Biotechnology – Writing a good paper on biotechnology is a challenging task. If you struggling to complete it, be sure to take a quick look at the following article.
You can find free research papers online as well. There are several documents that are available online. You can download them. You can check the image search as well if you are having trouble locating one. Try typing it into the search engine of the web browser and the image browser for the best results.
Video advice: HOW TO WRITE A RESEARCH PAPER
HOW TO WRITE A RESEARCH PAPER |Beginners Guide to Writing Quality Essays from An Oxford Grad Student

Guide for authors
Get more information about Current Research in Biotechnology. Check the Author information pack on Elsevier.com.
INTRODUCTION Current Research in Biotechnology is definitely an worldwide peer reviewed journal dedicated to publishing original research and short communications caused by research in Analytical biotechnology, Plant biotechnology, Food biotechnology, Energy biotechnology, Ecological biotechnology, Systems biology, Nanobiotechnology, Tissue, cell and path engineering, Chemical biotechnology, and Pharmaceutical biotechnology. The Journal publishes Research Papers, Short Communications, Graphical Reviews and Reviews. We offer the “Your Paper The Right Path” Elsevier guideline which enables authors to submit their primary manuscript file with no formatting needs. Research Papers aren’t limited in dimensions. However, we all do highly recommend to authors to become as succinct as you possibly can within the welfare from the readers and also the distribution from the work. Short Communications possess the following soft limits. The manuscript should ideally contain a maximum of 4-6 Figures/Tables and 4000 words, such as the title page, all parts of the manuscript (such as the references), and Figure/Table legends.
Structure for writing a scientific research proposal in biotechnology
The aim or goal and objective of the biotechnology research proposal should give a broad indication of the expected research outcome and the hypothesis to be tested can also be the aim of your study. The objective can be categorized as primary and secondary according to the parameters and tools used to achieve the goal.
Writing an investigation proposal in our era is definitely an entirely challenging mission due to the constant evolution within the research design and the necessity to incorporate innovative concepts and medical advances within the methodology section. A properly-formatted research proposal in the area of biotechnology is going to be written based on the needed guidelines forms the mainstay for that research, and therefore proposal writing is a vital step while performing research. The primary objective in preparing an investigation proposal would be to obtain approval from the 3 committees like the ethics committee and grant committee.
Research Papers – Learn more about research papers for the Master of Biotechnology Program at Northwestern University.
Alison Chow et al., “Metabolic engineering of the non-sporulating, non-solventogenic Clostridium acetobutylicum strain M5 to produce butanol without acetone demonstrate the robustness of the acid-formation pathways and the importance of the electron balance”, Metabolic Engineering 2008.
Natural Products and Biotechnology
Natural Products and Biotechnology (NatProBiotech) is an International Journal and only accepting English manuscripts. NatProBiotech publishes original research articles and review articles only.
- Research Article
- Review Article
Current Issue
Natural Products and Biotechnology (Nat. Pro. Biotech. ) (ISSN: 2791-674X) is an International Journal and only accepting English manuscripts. Natural Products and Biotechnology publishes original research articles and review articles only and publishes twice a year. There is no fee for article submission, article processing, or publication processes. Please write the article in good English. Choose only one of the British or American usage, you should not use both together. If the language of the article is not good enough, please have it edited by anEnglish Language Editing service. The article will be reviewed by the Spelling and Language editor, if the editor decides that it is not written in good English, your article will be send to corresponding author for edit before the referee process. Research articles should report the results of original research. The article should not have been published elsewhere. Review articles should cover current topics and comply with the journal’s publication guidelines and should not have been published anywhere before.
Video advice: How To Write Research Paper
How To Write Research Paper | Step-by-Step Research Paper Writing Process | by Aditi Sharma

What are the research topics in biotechnology?
- Research Areas.
- Cancer Biotechnology.
- Cardiovascular Biology & Transplantation Biology.
- Cell & Molecular Biology.
- Developmental Biology & Neurobiology.
- Diagnostics & Medical Devices.
- Drug Discovery & Delivery.
- Microbial & Environmental Biotechnology.
How do you publish a research paper in biotechnology?
How to publish your research paper in an international journal
- International journal of Environment, Agriculture and Biotechnology (IJEAB) publish research paper of related fields. ...
- Your research paper that you are going to submit should follow the same format that is mentioned in journal.
How do you research biotechnology?
Step-By-Step Guide To Becoming a Biotechnologist
- Step One: Earn a Bachelor's Degree (Four years) ...
- Step Two: Gain Practical Work Experience (Optional, Timeline Varies) ...
- Step Three: Earn a Certificate or Master's Degree In Biotechnology (One to Three Years)
How do you write a research paper step by step?
Basic Steps in the Research Process
- Step 1: Identify and develop your topic. ...
- Step 2 : Do a preliminary search for information. ...
- Step 3: Locate materials. ...
- Step 4: Evaluate your sources. ...
- Step 5: Make notes. ...
- Step 6: Write your paper. ...
- Step 7: Cite your sources properly. ...
- Step 8: Proofread.
What is biotechnology research?
Biotechnologists identify practical uses of biological material – including the physical, chemical, and genetic properties of cells – to improve agricultural, environmental, or pharmaceutical products, although biotechnologists also work in related capacities, as in marine biotechnology. ...
Related Articles:
- How To Write A Research Paper Biology
- How To Write Abstract For Science Research Paper
- How To Write Research Proposal For Phd In Biotechnology
- How To Write A Journal Paper In Engineering
- How To Write A Computer Science Paper
- How To Write A Review Paper In Chemistry
Science Journalist
Science atlas, our goal is to spark the curiosity that exists in all of us. We invite readers to visit us daily, explore topics of interest, and gain new perspectives along the way.
You may also like
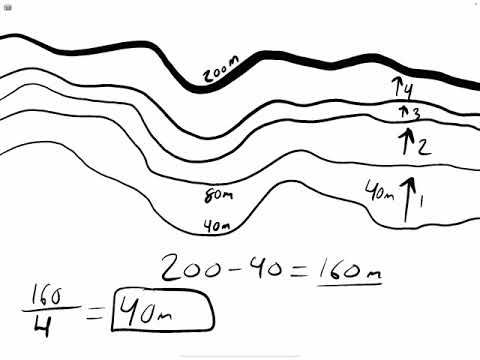
What Is A Contour Interval Geology

What Jobs Can I Do With A Science Degree

How To Use The Cricut Design Space App
Add comment, cancel reply.
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
Recent discoveries

How Is Nanotechnology Used In Electronics

Where Was Earth To Echo Filmed

What Is Metallurgy In Chemistry

What Is Future Directions For Science Fair
- Animals 3041
- Astronomy 8
- Biology 2281
- Chemistry 482
- Culture 1333
- Health 8466
- History 2152
- Physics 913
- Planet Earth 3239
- Science 2158
- Strange News 1230
- Technology 3625
Random fact
Worldwide Group of Scientists Reveal a brand new Kind of Botox treatment
Preparing your manuscript
This section provides general style and formatting information only. Formatting guidelines for specific article types can be found below.
- Research article
- Database article
- Software article
- Matters Arising
General formatting guidelines
- Preparing main manuscript text
- Preparing illustrations and figures
- Preparing tables
- Preparing additional files
Preparing figures
When preparing figures, please follow the formatting instructions below.
- Figures should be numbered in the order they are first mentioned in the text, and uploaded in this order. Multi-panel figures (those with parts a, b, c, d etc.) should be submitted as a single composite file that contains all parts of the figure.
- Figures should be uploaded in the correct orientation.
- Figure titles (max 15 words) and legends (max 300 words) should be provided in the main manuscript, not in the graphic file.
- Figure keys should be incorporated into the graphic, not into the legend of the figure.
- Each figure should be closely cropped to minimize the amount of white space surrounding the illustration. Cropping figures improves accuracy when placing the figure in combination with other elements when the accepted manuscript is prepared for publication on our site. For more information on individual figure file formats, see our detailed instructions.
- Individual figure files should not exceed 10 MB. If a suitable format is chosen, this file size is adequate for extremely high quality figures.
- Please note that it is the responsibility of the author(s) to obtain permission from the copyright holder to reproduce figures (or tables) that have previously been published elsewhere. In order for all figures to be open access, authors must have permission from the rights holder if they wish to include images that have been published elsewhere in non open access journals. Permission should be indicated in the figure legend, and the original source included in the reference list.
Figure file types
We accept the following file formats for figures:
- EPS (suitable for diagrams and/or images)
- PDF (suitable for diagrams and/or images)
- Microsoft Word (suitable for diagrams and/or images, figures must be a single page)
- PowerPoint (suitable for diagrams and/or images, figures must be a single page)
- TIFF (suitable for images)
- JPEG (suitable for photographic images, less suitable for graphical images)
- PNG (suitable for images)
- BMP (suitable for images)
- CDX (ChemDraw - suitable for molecular structures)
For information and suggestions of suitable file formats for specific figure types, please see our author academy .
Figure size and resolution
Figures are resized during publication of the final full text and PDF versions to conform to the BioMed Central standard dimensions, which are detailed below.
Figures on the web:
- width of 600 pixels (standard), 1200 pixels (high resolution).
Figures in the final PDF version:
- width of 85 mm for half page width figure
- width of 170 mm for full page width figure
- maximum height of 225 mm for figure and legend
- image resolution of approximately 300 dpi (dots per inch) at the final size
Figures should be designed such that all information, including text, is legible at these dimensions. All lines should be wider than 0.25 pt when constrained to standard figure widths. All fonts must be embedded.
Figure file compression
- Vector figures should if possible be submitted as PDF files, which are usually more compact than EPS files.
- TIFF files should be saved with LZW compression, which is lossless (decreases file size without decreasing quality) in order to minimize upload time.
- JPEG files should be saved at maximum quality.
- Conversion of images between file types (especially lossy formats such as JPEG) should be kept to a minimum to avoid degradation of quality.
If you have any questions or are experiencing a problem with figures, please contact the customer service team at [email protected] .
Preparing main manuscript text
Quick points:
- Use double line spacing
- Include line and page numbering
- Use SI units: Please ensure that all special characters used are embedded in the text, otherwise they will be lost during conversion to PDF
- Do not use page breaks in your manuscript
File formats
The following word processor file formats are acceptable for the main manuscript document:
- Microsoft word (DOC, DOCX)
- Rich text format (RTF)
- TeX/LaTeX
Please note: editable files are required for processing in production. If your manuscript contains any non-editable files (such as PDFs) you will be required to re-submit an editable file when you submit your revised manuscript, or after editorial acceptance in case no revision is necessary.
Additional information for TeX/LaTeX users
You are encouraged to use the Springer Nature LaTeX template when preparing a submission. A PDF of your manuscript files will be compiled during submission using pdfLaTeX and TexLive 2021. All relevant editable source files must be uploaded during the submission process. Failing to submit these source files will cause unnecessary delays in the production process.
Style and language
Improving your written english .
Presenting your work in well-written English gives it its best chance for editors and reviewers to understand it and evaluate it fairly.
We have some editing services that can help you to get your writing ready for submission.
Language quality checker
You can upload your manuscript and get a free language check from our partner AJE. The software uses AI to make suggestions that can improve writing quality. Trained on 300,000+ research manuscripts from more than 400+ areas of study and over 2000 field-specific topics the tool will deliver fast, highly accurate English language improvements. Your paper will be digitally edited and returned to you within approximately 10 minutes.
Try the tool for free now
Language and manuscript preparation services
Let one of our experts assist you with getting your manuscript and language into shape - our services cover:
- English language improvement
- scientific in-depth editing and strategic advice
- figure and tables formatting
- manuscript formatting to match your target journal
- specialist academic translation to English from Spanish, Portuguese, Japanese, or simplified Chinese
Get started and save 15%
Please note that using these tools, or any other service, is not a requirement for publication, nor does it imply or guarantee that editors will accept the article, or even select it for peer review.
您怎么做才有助于改进您的稿件以便顺利发表?
如果在结构精巧的稿件中用精心组织的英语展示您的作品,就能最大限度地让编辑和审稿人理解并公正评估您的作品。许多研究人员发现,获得一些独立支持有助于他们以尽可能美好的方式展示他们的成果。Springer Nature Author Services 的专家可帮助您准备稿件,具体 包括润色英语表述、添加有见地的注释、为稿件排版、设计图表、翻译等 。
开始使用即可节省 15% 的费用
您还可以使用我们的 免费语法检查工具 来评估您的作品。
请注意,使用这些工具或任何其他服务不是发表前必须满足的要求,也不暗示或保证相关文章定会被编辑接受(甚至未必会被选送同行评审)。
発表に備えて、論文を改善するにはどうすればよいでしょうか?
内容が適切に組み立てられ、質の高い英語で書かれた論文を投稿すれば、編集者や査読者が論文を理解し、公正に評価するための最善の機会となります。多くの研究者は、個別のサポートを受けることで、研究結果を可能な限り最高の形で発表できると思っています。Springer Nature Author Servicesのエキスパートが、 英文の編集、建設的な提言、論文の書式、図の調整、翻訳 など、論文の作成をサポートいたします。
今なら15%割引でご利用いただけます
原稿の評価に、無料 の文法チェック ツールもご利用いただけます。
これらのツールや他のサービスをご利用いただくことは、論文を掲載するための要件ではありません。また、編集者が論文を受理したり、査読に選定したりすることを示唆または保証するものではないことにご注意ください。
게재를 위해 원고를 개선하려면 어떻게 해야 할까요?
여러분의 작품을 체계적인 원고로 발표하는 것은 편집자와 심사자가 여러분의 연구를 이해하고 공정하게 평가할 수 있는 최선의 기회를 제공합니다. 많은 연구자들은 어느 정도 독립적인 지원을 받는 것이 가능한 한 최선의 방법으로 자신의 결과를 발표하는 데 도움이 된다고 합니다. Springer Nature Author Services 전문가들은 영어 편집, 발전적인 논평, 원고 서식 지정, 그림 준비, 번역 등과 같은 원고 준비를 도와드릴 수 있습니다.
지금 시작하면 15% 할인됩니다
또한 당사의 무료 문법 검사 도구를 사용하여 여러분의 연구를 평가할 수 있습니다.
이러한 도구 또는 기타 서비스를 사용하는 것은 게재를 위한 필수 요구사항이 아니며, 편집자가 해당 논문을 수락하거나 피어 리뷰에 해당 논문을 선택한다는 것을 암시하거나 보장하지는 않습니다.
¿Cómo puede ayudar a mejorar el artículo para su publicación?
Si presenta su trabajo en un artículo bien estructurado y en inglés bien escrito, los editores y revisores podrán comprenderlo mejor y evaluarlo de forma justa. Muchos investigadores piensan que un poco de apoyo independiente les ayuda a presentar los resultados de la mejor forma posible. Los expertos de Springer Nature Author Services pueden ayudarle a preparar el artículo con la edición en inglés, comentarios para su elaboración, el formato del artículo, la preparación de figuras, la traducción y mucho más.
Empiece ahora y ahorre un 15%
También puede usar nuestra herramienta gratuita Grammar Check para evaluar su trabajo.
Tenga en cuenta que utilizar estas herramientas, así como cualquier otro servicio, no es un requisito para publicación, y tampoco implica ni garantiza que los editores acepten el artículo, ni siquiera que lo seleccionen para revisión científica externa.
Como pode ajudar a melhorar o seu manuscrito para publicação?
Apresentar o seu trabalho num manuscrito bem estruturado e em inglês bem escrito confere-lhe a melhor probabilidade de os editores e revisores o compreenderem e avaliarem de forma justa. Muitos investigadores verificam que obter algum apoio independente os ajuda a apresentar os seus resultados da melhor forma possível. Os especialistas da Springer Nature Author Services podem ajudá-lo na preparação do manuscrito, incluindo edição de língua inglesa, comentários de desenvolvimento, formatação do manuscrito, preparação de figuras, tradução e muito mais.
Comece agora e poupe 15%
Também pode utilizar a nossa ferramenta gratuita de verificação de gramática para efetuar uma avaliação do seu trabalho.
Tenha em conta que a utilização destas ferramentas, ou de qualquer outro serviço, não constitui um requisito para publicação, nem implica nem garante que os editores aceitem o artigo ou o selecionem para revisão por pares.
Data and materials
For all journals, BioMed Central strongly encourages all datasets on which the conclusions of the manuscript rely to be either deposited in publicly available repositories (where available and appropriate) or presented in the main paper or additional supporting files, in machine-readable format (such as spread sheets rather than PDFs) whenever possible. Please see the list of recommended repositories in our editorial policies.
For some journals, deposition of the data on which the conclusions of the manuscript rely is an absolute requirement. Please check the Instructions for Authors for the relevant journal and article type for journal specific policies.
For all manuscripts, information about data availability should be detailed in an ‘Availability of data and materials’ section. For more information on the content of this section, please see the Declarations section of the relevant journal’s Instruction for Authors. For more information on BioMed Centrals policies on data availability, please see our [editorial policies].
Formatting the 'Availability of data and materials' section of your manuscript
The following format for the 'Availability of data and materials section of your manuscript should be used:
"The dataset(s) supporting the conclusions of this article is(are) available in the [repository name] repository, [unique persistent identifier and hyperlink to dataset(s) in http:// format]."
The following format is required when data are included as additional files:
"The dataset(s) supporting the conclusions of this article is(are) included within the article (and its additional file(s))."
BioMed Central endorses the Force 11 Data Citation Principles and requires that all publicly available datasets be fully referenced in the reference list with an accession number or unique identifier such as a DOI.
For databases, this section should state the web/ftp address at which the database is available and any restrictions to its use by non-academics.
For software, this section should include:
- Project name: e.g. My bioinformatics project
- Project home page: e.g. http://sourceforge.net/projects/mged
- Archived version: DOI or unique identifier of archived software or code in repository (e.g. enodo)
- Operating system(s): e.g. Platform independent
- Programming language: e.g. Java
- Other requirements: e.g. Java 1.3.1 or higher, Tomcat 4.0 or higher
- License: e.g. GNU GPL, FreeBSD etc.
- Any restrictions to use by non-academics: e.g. licence needed
Information on available repositories for other types of scientific data, including clinical data, can be found in our editorial policies .
See our editorial policies for author guidance on good citation practice.
Please check the submission guidelines for the relevant journal and article type.
What should be cited?
Only articles, clinical trial registration records and abstracts that have been published or are in press, or are available through public e-print/preprint servers, may be cited.
Unpublished abstracts, unpublished data and personal communications should not be included in the reference list, but may be included in the text and referred to as "unpublished observations" or "personal communications" giving the names of the involved researchers. Obtaining permission to quote personal communications and unpublished data from the cited colleagues is the responsibility of the author. Only footnotes are permitted. Journal abbreviations follow Index Medicus/MEDLINE.
Any in press articles cited within the references and necessary for the reviewers' assessment of the manuscript should be made available if requested by the editorial office.
How to format your references
Please check the Instructions for Authors for the relevant journal and article type for examples of the relevant reference style.
Web links and URLs: All web links and URLs, including links to the authors' own websites, should be given a reference number and included in the reference list rather than within the text of the manuscript. They should be provided in full, including both the title of the site and the URL, as well as the date the site was accessed, in the following format: The Mouse Tumor Biology Database. http://tumor.informatics.jax.org/mtbwi/index.do . Accessed 20 May 2013. If an author or group of authors can clearly be associated with a web link, such as for weblogs, then they should be included in the reference.
Authors may wish to make use of reference management software to ensure that reference lists are correctly formatted.
Preparing tables
When preparing tables, please follow the formatting instructions below.
- Tables should be numbered and cited in the text in sequence using Arabic numerals (i.e. Table 1, Table 2 etc.).
- Tables less than one A4 or Letter page in length can be placed in the appropriate location within the manuscript.
- Tables larger than one A4 or Letter page in length can be placed at the end of the document text file. Please cite and indicate where the table should appear at the relevant location in the text file so that the table can be added in the correct place during production.
- Larger datasets, or tables too wide for A4 or Letter landscape page can be uploaded as additional files. Please see [below] for more information.
- Tabular data provided as additional files can be uploaded as an Excel spreadsheet (.xls ) or comma separated values (.csv). Please use the standard file extensions.
- Table titles (max 15 words) should be included above the table, and legends (max 300 words) should be included underneath the table.
- Tables should not be embedded as figures or spreadsheet files, but should be formatted using ‘Table object’ function in your word processing program.
- Color and shading may not be used. Parts of the table can be highlighted using superscript, numbering, lettering, symbols or bold text, the meaning of which should be explained in a table legend.
- Commas should not be used to indicate numerical values.
If you have any questions or are experiencing a problem with tables, please contact the customer service team at [email protected] .
Preparing additional files
As the length and quantity of data is not restricted for many article types, authors can provide datasets, tables, movies, or other information as additional files.
All Additional files will be published along with the accepted article. Do not include files such as patient consent forms, certificates of language editing, or revised versions of the main manuscript document with tracked changes. Such files, if requested, should be sent by email to the journal’s editorial email address, quoting the manuscript reference number. Please do not send completed patient consent forms unless requested.
Results that would otherwise be indicated as "data not shown" should be included as additional files. Since many web links and URLs rapidly become broken, BioMed Central requires that supporting data are included as additional files, or deposited in a recognized repository. Please do not link to data on a personal/departmental website. Do not include any individual participant details. The maximum file size for additional files is 20 MB each, and files will be virus-scanned on submission. Each additional file should be cited in sequence within the main body of text.
If additional material is provided, please list the following information in a separate section of the manuscript text:
- File name (e.g. Additional file 1)
- File format including the correct file extension for example .pdf, .xls, .txt, .pptx (including name and a URL of an appropriate viewer if format is unusual)
- Title of data
- Description of data
Additional files should be named "Additional file 1" and so on and should be referenced explicitly by file name within the body of the article, e.g. 'An additional movie file shows this in more detail [see Additional file 1]'.
For further guidance on how to use Additional files or recommendations on how to present particular types of data or information, please see How to use additional files .
Important information
Editorial board
For authors
For editorial board members
For reviewers
- Manuscript editing services
- Follow us on Twitter
Annual Journal Metrics
2022 Citation Impact 3.5 - 2-year Impact Factor 3.5 - 5-year Impact Factor 0.880 - SNIP (Source Normalized Impact per Paper) 0.654 - SJR (SCImago Journal Rank)
2023 Speed 10 days submission to first editorial decision for all manuscripts (Median) 155 days submission to accept (Median)
2023 Usage 1,134,875 downloads 518 Altmetric mentions
- More about our metrics
BMC Biotechnology
ISSN: 1472-6750
- General enquiries: [email protected]
Browse Course Material
Course info, instructors.
- Prof. Christopher Burge
- Prof. David Sabatini
- Dr. Marilee Ogren-Balkema
- Dr. Alice Rushforth
Departments
As taught in.
- Biotechnology
- Molecular Biology
Learning Resource Types
Experimental molecular biology: biotechnology ii, scientific comm..
This course includes significant instruction in scientific communications. During the term, Dr. Marilee Ogren-Balkema presents ten lectures on a range of reading, presentation and writing topics.
Background reading
Gopen, George D., and Judith A. Swan. “ The Science of Scientific Writing .” The American Scientist 78 (1990): 550-558.
Lectures on Scientific Communications
1: Basic Scientific Communication ( PDF )
2: How to Review the Literature ( PDF )
3: How To Write a Research Proposal ( PDF )
4: Preparing Effective Oral Presentations ( PDF )
5: How to Write a Mini Literature Review ( PDF )
6: How to Write a Research Paper I: Illustrations ( PDF - 1.2 MB )
7: How to Write a Research Paper II: Results Section ( PDF )
8: How to Write a Research Paper III: Methods Section ( PDF )
9: How to Write a Research Paper IV: Introduction and Discussion ( PDF )
10: How to Write a Research Paper V: Title and Abstract ( PDF )

You are leaving MIT OpenCourseWare

- Collections
How to Write a Research Proposal in Biotechnology
This comprehensive guide will break down the process into easy-to-follow steps, taking you from writing a research proposal with a focused question to outlining a clear research proposal structure . We’ll delve into each key section, including the literature review for your research proposal and the most appropriate methodology for your study. By the end, you’ll be equipped to write a compelling research proposal that sets your biotechnology project on the path to success.
Subject of Study
- Clearly define the specific area of biotechnology you’ll be exploring.
- For example, are you researching biofuels, gene editing, bioremediation, or a specific disease application?
- Narrow your focus to a manageable topic within your chosen area.
Research Question
- Craft a concise and focused question (under 15 words) that your research aims to answer.
- This question should be clear, relevant, and feasible within the scope of your project.
Analysis of the Problem
- Briefly introduce your chosen topic and its significance in the field of biotechnology.
- Explain the problem or gap in knowledge you’re addressing.
- Consider including a concept map to visually represent the relationships between key factors in your study.
- This section should be between 75-300 words.
Research Method
- Describe the specific research methods you plan to use.
- Experiments (e.g., cell culture, protein purification)
- Computational modeling
- Surveys of relevant specialists
- Literature reviews and meta-analyses of existing data
- Justify why your chosen methods are the most appropriate for answering your research question.
- Explain how you’ll ensure the validity and reliability of your data collection process. (This section is also 75-300 words)
Preliminary Answer to the Research Question
- Based on your initial understanding and literature review, what do you expect to find?
- Briefly summarize your preliminary hypothesis or expected outcomes.
- This is not a definitive answer, but rather an educated guess based on current knowledge. (75-300 words)
Academic Contribution of the Study
- Explain how your research will contribute to the field of biotechnology.
- Will it fill a gap in knowledge, propose a new theory, or develop a novel application?
- Highlight the potential impact of your findings on existing theories, practices, or policies within biotechnology. (75-300 words)
Literature Review
- Dedicating more space (up to 300 words) allows for a more in-depth review.
- Discuss key studies, theories, and methodologies relevant to your research question.
- Critically analyze existing literature, identifying strengths, weaknesses, and any unanswered questions that your research aims to address.
- Provide a concise bibliography of the main academic sources used in your study area.
Trends in biotechnology

IBM RXN for Chemistry AI | Computational chemistry AI
Machine learning models for genomic classification and prediction, 3d bioprinting: 3d bioprinting for tissue and organ regeneration, trends in biotechnology: lab-grown meats, trends in biotechnology: molecular jackhammers, importance of research proposals.
Research proposals serve several important purposes and are required in a variety of settings:
Securing Funding: A well-written proposal is essential for convincing granting agencies, institutions, or private donors to fund your research. It showcases the significance of your project, your research plan, and your qualifications to carry it out.
Planning and Development: The process of crafting a proposal forces you to clearly define your research question, methodology, and expected outcomes. This clarity helps you refine your research focus, identify potential roadblocks, and ensure your project is feasible within your timeframe and resources.
Communication and Review: A proposal clearly communicates your research goals and methods to reviewers and collaborators. This allows them to assess the merit of your project, offer feedback, and identify potential areas for improvement.
Here are some specific contexts where research proposals are commonly required:
- Academic Research: Master’s and doctoral students typically need to submit proposals for their thesis or dissertation research.
- Grant Applications: Researchers seeking funding from government agencies, private foundations, or universities often require a detailed proposal outlining their project.
- Industry Research and Development: Companies involved in research and development may require proposals for new projects or collaborations.
- Ethics Review Boards: Research involving human subjects or sensitive data may require approval from an ethics review board, which often involves submitting a proposal outlining the research plan and potential risks.
Overall, a research proposal is a vital tool for researchers to secure funding, refine their research plan, and gain approval for their projects.
Sign Up For Daily Newsletter
Be keep up get the latest articles delivered straight to your inbox..
I have read and agree to the terms & conditions
Leave a review Cancel reply
Your email address will not be published. Required fields are marked *
Your comment *
Your name *
Your Email *
Biotechtutorials
Share knowledge..
- Quick Links

Username or Email Address
Remember Me
Biotechnology Research Paper Topics

This collection of biotechnology research paper topics provides the list of 10 potential topics for research papers and overviews the history of biotechnology.

Academic Writing, Editing, Proofreading, And Problem Solving Services
Get 10% off with 24start discount code, 1. animal breeding: genetic methods.
Modern animal breeding relies on scientific methods to control production of domesticated animals, both livestock and pets, which exhibit desired physical and behavioral traits. Genetic technology aids animal breeders to attain nutritional, medical, recreational, and fashion standards demanded by consumers for animal products including meat, milk, eggs, leather, wool, and pharmaceuticals. Animals are also genetically designed to meet labor and sporting requirements for speed and endurance, conformation and beauty ideals to win show competitions, and intelligence levels to perform obediently at tasks such as herding, hunting, and tracking. By the late twentieth century, genetics and mathematical models were appropriated to identify the potential of immature animals. DNA markers indicate how young animals will mature, saving breeders money by not investing in animals lacking genetic promise. Scientists also successfully transplanted sperm-producing stem cells with the goal of restoring fertility to barren breeding animals. At the National Animal Disease Center in Ames, Iowa, researchers created a gene-based test, which uses a cloned gene of the organism that causes Johne’s disease in cattle in order to detect that disease to avert epidemics. Researchers also began mapping the dog genome and developing molecular techniques to evaluate canine chromosomes in the Quantitative Trait Loci (QTL). Bioinformatics incorporates computers to analyze genetic material. Some tests were developed to diagnose many of several hundred genetic canine diseases including hip dysplasia and progressive retinal atrophy (PRA). A few breed organizations modified standards to discourage breeding of genetically flawed animals and promote heterozygosity.
2. Antibacterial Chemotherapy
In the early years of the twentieth century, the search for agents that would be effective against internal infections proceeded along two main routes. The first was a search for naturally occurring substances that were effective against microorganisms (antibiosis). The second was a search for chemicals that would have the same effect (chemotherapy). Despite the success of penicillin in the 1940s, the major early advances in the treatment of infection occurred not through antibiosis but through chemotherapy. The principle behind chemotherapy was that there was a relationship between chemical structure and pharmacological action. The founder of this concept was Paul Erhlich (1854–1915). An early success came in 1905 when atoxyl (an organic arsenic compound) was shown to destroy trypanosomes, the microbes that caused sleeping sickness. Unfortunately, atoxyl also damaged the optic nerve. Subsequently, Erhlich and his co-workers synthesized and tested hundreds of related arsenic compounds. Ehrlich was a co-recipient (with Ilya Ilyich Mechnikov) of the Nobel Prize in medicine in 1908 for his work on immunity. Success in discovering a range of effective antibacterial drugs had three important consequences: it brought a range of important diseases under control for the first time; it provided a tremendous stimulus to research workers and opened up new avenues of research; and in the resulting commercial optimism, it led to heavy postwar investment in the pharmaceutical industry. The therapeutic revolution had begun.

3. Artificial Insemination and in Vitro Fertilization
Artificial insemination (AI) involves the extraction and collection of semen together with techniques for depositing semen in the uterus in order to achieve successful fertilization and pregnancy. Throughout the twentieth century, the approach has offered animal breeders the advantage of being able to utilize the best available breeding stock and at the correct time within the female reproductive cycle, but without the limitations of having the animals in the same location. AI has been applied most intensively within the dairy and beef cattle industries and to a lesser extent horse breeding and numerous other domesticated species.
Many of the techniques involved in artificial insemination would lay the foundation for in vitro fertilization (IVF) in the latter half of the twentieth century. IVF refers to the group of technologies that allow fertilization to take place outside the body involving the retrieval of ova or eggs from the female and sperm from the male, which are then combined in artificial, or ‘‘test tube,’’ conditions leading to fertilization. The fertilized eggs then continue to develop for several days ‘‘in culture’’ until being transferred to the female recipient to continue developing within the uterus.
4. Biopolymers
Biopolymers are natural polymers, long-chained molecules (macromolecules) consisting mostly of a repeated composition of building blocks or monomers that are formed and utilized by living organisms. Each group of biopolymers is composed of different building blocks, for example chains of sugar molecules form starch (a polysaccharide), chains of amino acids form proteins and peptides, and chains of nucleic acid form DNA and RNA (polynucleotides). Biopolymers can form gels, fibers, coatings, and films depending on the specific polymer, and serve a variety of critical functions for cells and organisms. Proteins including collagens, keratins, silks, tubulins, and actin usually form structural composites or scaffolding, or protective materials in biological systems (e.g., spider silk). Polysaccharides function in molecular recognition at cell membrane surfaces, form capsular barrier layers around cells, act as emulsifiers and adhesives, and serve as skeletal or architectural materials in plants. In many cases these polymers occur in combination with proteins to form novel composite structures such as invertebrate exoskeletons or microbial cell walls, or with lignin in the case of plant cell walls.
The use of the word ‘‘cloning’’ is fraught with confusion and inconsistency, and it is important at the outset of this discussion to offer definitional clarification. For instance, in the 1997 article by Ian Wilmut and colleagues announcing the birth of the first cloned adult vertebrate (a ewe, Dolly the sheep) from somatic cell nuclear transfer, the word clone or cloning was never used, and yet the announcement raised considerable disquiet about the prospect of cloned human beings. In a desire to avoid potentially negative forms of language, many prefer to substitute ‘‘cell expansion techniques’’ or ‘‘therapeutic cloning’’ for cloning. Cloning has been known for centuries as a horticultural propagation method: for example, plants multiplied by grafting, budding, or cuttings do not differ genetically from the original plant. The term clone entered more common usage as a result of a speech in 1963 by J.B.S. Haldane based on his paper, ‘‘Biological possibilities for the human species of the next ten-thousand years.’’ Notwithstanding these notes of caution, we can refer to a number of processes as cloning. At the close of the twentieth century, such techniques had not yet progressed to the ability to bring a cloned human to full development; however, the ability to clone cells from an adult human has potential to treat diseases. International policymaking in the late 1990s sought to distinguish between the different end uses for somatic cell nuclear transfer resulting in the widespread adoption of the distinction between ‘‘reproductive’’ and ‘‘therapeutic’’ cloning. The function of the distinction has been to permit the use (in some countries) of the technique to generate potentially beneficial therapeutic applications from embryonic stem cell technology whilst prohibiting its use in human reproduction. In therapeutic applications, nuclear transfer from a patient’s cells into an enucleated ovum is used to create genetically identical embryos that would be grown in vitro but not be allowed to continue developing to become a human being. The resulting cloned embryos could be used as a source from which to produce stem cells that can then be induced to specialize into the specific type of tissue required by the patient (such as skin for burns victims, brain neuron cells for Parkinson’s disease sufferers, or pancreatic cells for diabetics). The rationale is that because the original nuclear material is derived from a patient’s adult tissue, the risks of rejection of such cells by the immune system are reduced.
6. Gene Therapy
In 1971, Australian Nobel laureate Sir F. MacFarlane Burnet thought that gene therapy (introducing genes into body tissue, usually to treat an inherited genetic disorder) looked more and more like a case of the emperor’s new clothes. Ethical issues aside, he believed that practical considerations forestalled possibilities for any beneficial gene strategy, then or probably ever. Bluntly, he wrote: ‘‘little further advance can be expected from laboratory science in the handling of ‘intrinsic’ types of disability and disease.’’ Joshua Lederberg and Edward Tatum, 1958 Nobel laureates, theorized in the 1960s that genes might be altered or replaced using viral vectors to treat human diseases. Stanfield Rogers, working from the Oak Ridge National Laboratory in 1970, had tried but failed to cure argininemia (a genetic disorder of the urea cycle that causes neurological damage in the form of mental retardation, seizures, and eventually death) in two German girls using Swope papilloma virus. Martin Cline at the University of California in Los Angeles, made the second failed attempt a decade later. He tried to correct the bone marrow cells of two beta-thalassemia patients, one in Israel and the other in Italy. What Cline’s failure revealed, however, was that many researchers who condemned his trial as unethical were by then working toward similar goals and targeting different diseases with various delivery methods. While Burnet’s pessimism finally proved to be wrong, progress in gene therapy was much slower than antibiotic or anticancer chemotherapy developments over the same period of time. While gene therapy had limited success, it nevertheless remained an active area for research, particularly because the Human Genome Project, begun in 1990, had resulted in a ‘‘rough draft’’ of all human genes by 2001, and was completed in 2003. Gene mapping created the means for analyzing the expression patterns of hundreds of genes involved in biological pathways and for identifying single nucleotide polymorphisms (SNPs) that have diagnostic and therapeutic potential for treating specific diseases in individuals. In the future, gene therapies may prove effective at protecting patients from adverse drug reactions or changing the biochemical nature of a person’s disease. They may also target blood vessel formation in order to prevent heart disease or blindness due to macular degeneration or diabetic retinopathy. One of the oldest ideas for use of gene therapy is to produce anticancer vaccines. One method involves inserting a granulocyte-macrophage colony-stimulating factor gene into prostate tumor cells removed in surgery. The cells then are irradiated to prevent any further cancer and injected back into the same patient to initiate an immune response against any remaining metastases. Whether or not such developments become a major treatment modality, no one now believes, as MacFarland Burnet did in 1970, that gene therapy science has reached an end in its potential to advance health.
7. Genetic Engineering
The term ‘‘genetic engineering’’ describes molecular biology techniques that allow geneticists to analyze and manipulate deoxyribonucleic acid (DNA). At the close of the twentieth century, genetic engineering promised to revolutionize many industries, including microbial biotechnology, agriculture, and medicine. It also sparked controversy over potential health and ecological hazards due to the unprecedented ability to bypass traditional biological reproduction.
For centuries, if not millennia, techniques have been employed to alter the genetic characteristics of animals and plants to enhance specifically desired traits. In a great many cases, breeds with which we are most familiar bear little resemblance to the wild varieties from which they are derived. Canine breeds, for instance, have been selectively tailored to changing esthetic tastes over many years, altering their appearance, behavior and temperament. Many of the species used in farming reflect long-term alterations to enhance meat, milk, and fleece yields. Likewise, in the case of agricultural varieties, hybridization and selective breeding have resulted in crops that are adapted to specific production conditions and regional demands. Genetic engineering differs from these traditional methods of plant and animal breeding in some very important respects. First, genes from one organism can be extracted and recombined with those of another (using recombinant DNA, or rDNA, technology) without either organism having to be of the same species. Second, removing the requirement for species reproductive compatibility, new genetic combinations can be produced in a much more highly accelerated way than before. Since the development of the first rDNA organism by Stanley Cohen and Herbert Boyer in 1973, a number of techniques have been found to produce highly novel products derived from transgenic plants and animals.
At the same time, there has been an ongoing and ferocious political debate over the environmental and health risks to humans of genetically altered species. The rise of genetic engineering may be characterized by developments during the last three decades of the twentieth century.
8. Genetic Screening and Testing
The menu of genetic screening and testing technologies now available in most developed countries increased rapidly in the closing years of the twentieth century. These technologies emerged within the context of rapidly changing social and legal contexts with regard to the medicalization of pregnancy and birth and the legalization of abortion. The earliest genetic screening tests detected inborn errors of metabolism and sex-linked disorders. Technological innovations in genomic mapping and DNA sequencing, together with an explosion in research on the genetic basis of disease which culminated in the Human Genome Project (HGP), led to a range of genetic screening and testing for diseases traditionally recognized as genetic in origin and for susceptibility to more common diseases such as certain types of familial cancer, cardiac conditions, and neurological disorders among others. Tests were also useful for forensic, or nonmedical, purposes. Genetic screening techniques are now available in conjunction with in vitro fertilization and other types of reproductive technologies, allowing the screening of fertilized embryos for certain genetic mutations before selection for implantation. At present selection is purely on disease grounds and selection for other traits (e.g., for eye or hair color, intelligence, height) cannot yet be done, though there are concerns for eugenics and ‘‘designer babies.’’ Screening is available for an increasing number of metabolic diseases through tandem mass spectrometry, which uses less blood per test, allows testing for many conditions simultaneously, and has a very low false-positive rate as compared to conventional Guthrie testing. Finally, genetic technologies are being used in the judicial domain for determination of paternity, often associated with child support claims, and for forensic purposes in cases where DNA material is available for testing.
9. Plant Breeding: Genetic Methods
The cultivation of plants is the world’s oldest biotechnology. We have continually tried to produce improved varieties while increasing yield, features to aid cultivation and harvesting, disease, and pest resistance, or crop qualities such as longer postharvest storage life and improved taste or nutritional value. Early changes resulted from random crosspollination, rudimentary grafting, or spontaneous genetic change. For centuries, man kept the seed from the plants with improved characteristics to plant the following season’s crop. The pioneering work of Gregor Mendel and his development of the basic laws of heredity showed for other first time that some of the processes of heredity could be altered by experimental means. The genetic analysis of bacterial (prokaryote) genes and techniques for analysis of the higher (eukaryotic) organisms such as plants developed in parallel streams, but the rediscovery of Mendel’s work in 1900 fueled a burst of activity on understanding the role of genes in inheritance. The knowledge that genes are linked along the chromosome thereby allowed mapping of genes (transduction analysis, conjugation analysis, and transformation analysis). The power of genetics to produce a desirable plant was established, and it was appreciated that controlled breeding (test crosses and back crosses) and careful analysis of the progeny could distinguish traits that were dominant or recessive, and establish pure breeding lines. Traditional horticultural techniques of artificial self-pollination and cross-pollination were also used to produce hybrids. In the 1930s the Russian Nikolai Vavilov recognized the value of genetic diversity in domesticated crop plants and their wild relatives to crop improvement, and collected seeds from the wild to study total genetic diversity and use these in breeding programs. The impact of scientific crop breeding was established by the ‘‘Green revolution’’ of the 1960s, when new wheat varieties with higher yields were developed by careful crop breeding. ‘‘Mutation breeding’’— inducing mutations by exposing seeds to x-rays or chemicals such as sodium azide, accelerated after World War II. It was also discovered that plant cells and tissues grown in tissue culture would mutate rapidly. In the 1970s, haploid breeding, which involves producing plants from two identical sets of chromosomes, was extensively used to create new cultivars. In the twenty-first century, haploid breeding could speed up plant breeding by shortening the breeding cycle.
10. Tissue Culturing
The technique of tissue or cell culture, which relates to the growth of tissue or cells within a laboratory setting, underlies a phenomenal proportion of biomedical research. Though it has roots in the late nineteenth century, when numerous scientists tried to grow samples in alien environments, cell culture is credited as truly beginning with the first concrete evidence of successful growth in vitro, demonstrated by Johns Hopkins University embryologist Ross Harrison in 1907. Harrison took sections of spinal cord from a frog embryo, placed them on a glass cover slip and bathed the tissue in a nutrient media. The results of the experiment were startling—for the first time scientists visualized actual nerve growth as it would happen in a living organism—and many other scientists across the U.S. and Europe took up culture techniques. Rather unwittingly, for he was merely trying to settle a professional dispute regarding the origin of nerve fibers, Harrison fashioned a research tool that has since been designated by many as the greatest advance in medical science since the invention of the microscope.
From the 1980s, cell culture has once again been brought to the forefront of cancer research in the isolation and identification of numerous cancer causing oncogenes. In addition, cell culturing continues to play a crucial role in fields such as cytology, embryology, radiology, and molecular genetics. In the future, its relevance to direct clinical treatment might be further increased by the growth in culture of stem cells and tissue replacement therapies that can be tailored for a particular individual. Indeed, as cell culture approaches its centenary, it appears that its importance to scientific, medical, and commercial research the world over will only increase in the twenty-first century.
History of Biotechnology
Biotechnology grew out of the technology of fermentation, which was called zymotechnology. This was different from the ancient craft of brewing because of its thought-out relationships to science. These were most famously conceptualized by the Prussian chemist Georg Ernst Stahl (1659–1734) in his 1697 treatise Zymotechnia Fundamentalis, in which he introduced the term zymotechnology. Carl Balling, long-serving professor in Prague, the world center of brewing, drew on the work of Stahl when he published his Bericht uber die Fortschritte der zymotechnische Wissenschaften und Gewerbe (Account of the Progress of the Zymotechnic Sciences and Arts) in the mid-nineteenth century. He used the idea of zymotechnics to compete with his German contemporary Justus Liebig for whom chemistry was the underpinning of all processes.
By the end of the nineteenth century, there were attempts to develop a new scientific study of fermentation. It was an aspect of the ‘‘second’’ Industrial Revolution during the period from 1870 to 1914. The emergence of the chemical industry is widely taken as emblematic of the formal research and development taking place at the time. The development of microbiological industries is another example. For the first time, Louis Pasteur’s germ theory made it possible to provide convincing explanations of brewing and other fermentation processes.
Pasteur had published on brewing in the wake of France’s humiliation in the Franco–Prussian war (1870–1871) to assert his country’s superiority in an industry traditionally associated with Germany. Yet the science and technology of fermentation had a wide range of applications including the manufacture of foods (cheese, yogurt, wine, vinegar, and tea), of commodities (tobacco and leather), and of chemicals (lactic acid, citric acid, and the enzyme takaminase). The concept of zymotechnology associated principally with the brewing of beer began to appear too limited to its principal exponents. At the time, Denmark was the world leader in creating high-value agricultural produce. Cooperative farms pioneered intensive pig fattening as well as the mass production of bacon, butter, and beer. It was here that the systems of science and technology were integrated and reintegrated, conceptualized and reconceptualized.
The Dane Emil Christian Hansen discovered that infection from wild yeasts was responsible for numerous failed brews. His contemporary Alfred Jørgensen, a Copenhagen consultant closely associated with the Tuborg brewery, published a widely used textbook on zymotechnology. Microorganisms and Fermentation first appeared in Danish 1889 and would be translated, reedited, and reissued for the next 60 years.
The scarcity of resources on both sides during World War I brought together science and technology, further development of zymotechnology, and formulation of the concept of biotechnology. Impending and then actual war accelerated the use of fermentation technologies to make strategic materials. In Britain a variant of a process to ferment starch to make butadiene for synthetic rubber production was adapted to make acetone needed in the manufacture of explosives. The process was technically important as the first industrial sterile fermentation and was strategically important for munitions supplies. The developer, chemist Chaim Weizmann, later became well known as the first president of Israel in 1949.
In Germany scarce oil-based lubricants were replaced by glycerol made by fermentation. Animal feed was derived from yeast grown with the aid of the new synthetic ammonia in another wartime development that inspired the coining of the word biotechnology. Hungary was the agricultural base of the Austro–Hungarian empire and aspired to Danish levels of efficiency. The economist Karl Ereky (1878–1952) planned to go further and build the largest industrial pig-processing factory. He envisioned a site that would fatten 50,000 swine at a time while railroad cars of sugar beet arrived and fat, hides, and meat departed. In this forerunner of the Soviet collective farm, peasants (in any case now falling prey to the temptations of urban society) would be completely superseded by the industrialization of the biological process in large factory-like animal processing units. Ereky went further in his ruminations over the meaning of his innovation. He suggested that it presaged an industrial revolution that would follow the transformation of chemical technology. In his book entitled Biotechnologie, he linked specific technical injunctions to wide-ranging philosophy. Ereky was neither isolated nor obscure. He had been trained in the mainstream of reflection on the meaning of the applied sciences in Hungary, which would be remarkably productive across the sciences. After World War I, Ereky served as Hungary’s minister of food in the short-lived right wing regime that succeeded the fall of the communist government of Bela Kun.
Nonetheless it was not through Ereky’s direct action that his ideas seem to have spread. Rather, his book was reviewed by the influential Paul Lindner, head of botany at the Institut fu¨ r Ga¨ rungsgewerbe in Berlin, who suggested that microorganisms could also be seen as biotechnological machines. This concept was already found in the production of yeast and in Weizmann’s work with strategic materials, which was widely publicized at that very time. It was with this meaning that the word ‘‘Biotechnologie’’ entered German dictionaries in the 1920s.
Biotechnology represented more than the manipulation of existing organisms. From the beginning it was concerned with their improvement as well, and this meant the enhancement of all living creatures. Most dramatically this would include humanity itself; more mundanely it would include plants and animals of agricultural importance. The enhancement of people was called eugenics by the Victorian polymath and cousin of Charles Darwin, Francis Galton. Two strains of eugenics emerged: negative eugenics associated with weeding out the weak and positive eugenics associated with enhancing strength. In the early twentieth century, many eugenics proponents believed that the weak could be made strong. People had after all progressed beyond their biological limits by means of technology.
Jean-Jacques Virey, a follower of the French naturalist Jean-Baptiste de Monet de Lamarck, had coined the term ‘‘biotechnie’’ in 1828 to describe man’s ability to make technology do the work of biology, but it was not till a century later that the term entered widespread use. The Scottish biologist and town planner Patrick Geddes made biotechnics popular in the English-speaking world. Geddes, too, sought to link life and technology. Before World War I he had characterized the technological evolution of mankind as a move from the paleotechnic era of coal and iron to the neotechnic era of chemicals, electricity, and steel. After the war, he detected a new era based on biology—the biotechnic era. Through his friend, writer Lewis Mumford, Geddes would have great influence. Mumford’s book Technics and Civilization, itself a founding volume of the modern historiography of technology, promoted his vision of the Geddesian evolution.
A younger generation of English experimental biologists with a special interest in genetics, including J. B. S. Haldane, Julian Huxley, and Lancelot Hogben, also promoted a concept of biotechnology in the period between the world wars. Because they wrote popular works, they were among Britain’s best-known scientists. Haldane wrote about biological invention in his far-seeing work Daedalus. Huxley looked forward to a blend of social and eugenics-based biological engineering. Hogben, following Geddes, was more interested in engineering plants through breeding. He tied the progressivism of biology to the advance of socialism.
The improvement of the human race, genetic manipulation of bacteria, and the development of fermentation technology were brought together by the development of penicillin during World War II. This drug was successfully extracted from the juice exuded by a strain of the Penicillium fungus. Although discovered by accident and then developed further for purely scientific reasons, the scarce and unstable ‘‘antibiotic’’ called penicillin was transformed during World War II into a powerful and widely used drug. Large networks of academic and government laboratories and pharmaceutical manufacturers in Britain and the U.S. were coordinated by agencies of the two governments. An unanticipated combination of genetics, biochemistry, chemistry, and chemical engineering skills had been required. When the natural mold was bombarded with high-frequency radiation, far more productive mutants were produced, and subsequently all the medicine was made using the product of these man-made cells. By the 1950s penicillin was cheap to produce and globally available.
The new technology of cultivating and processing large quantities of microorganisms led to calls for a new scientific discipline. Biochemical engineering was one term, and applied microbiology another. The Swedish biologist, Carl-Goran Heden, possibly influenced by German precedents, favored the term ‘‘Biotechnologi’’ and persuaded his friend Elmer Gaden to relabel his new journal Biotechnology and Biochemical Engineering. From 1962 major international conferences were held under the banner of the Global Impact of Applied Microbiology. During the 1960s food based on single-cell protein grown in fermenters on oil or glucose seemed, to visionary engineers and microbiologists and to major companies, to offer an immediate solution to world hunger. Tropical countries rich in biomass that could be used as raw material for fermentation were also the world’s poorest. Alcohol could be manufactured by fermenting such starch or sugar rich crops as sugar cane and corn. Brazil introduced a national program of replacing oil-based petrol with alcohol in the 1970s.
It was not, however, just the developing countries that hoped to benefit. The Soviet Union developed fermentation-based protein as a major source of animal feed through the 1980s. In the U.S. it seemed that oil from surplus corn would solve the problem of low farm prices aggravated by the country’s boycott of the USSR in1979, and the term ‘‘gasohol‘‘ came into currency. Above all, the decline of established industries made the discovery of a new wealth maker an urgent priority for Western governments. Policy makers in both Germany and Japan during the 1970s were driven by a sense of the inadequacy of the last generation of technologies. These were apparently maturing, and the succession was far from clear. Even if electronics or space travel offered routes to the bright industrial future, these fields seemed to be dominated by the U.S. Seeing incipient crisis, the Green, or environmental, movement promoted a technology that would depend on renewable resources and on low-energy processes that would produce biodegradable products, recycle waste, and address problems of the health and nutrition of the world.
In 1973 the German government, seeking a new and ‘‘greener’’ industrial policy, commissioned a report entitled Biotechnologie that identified ways in which biological processing was key to modern developments in technology. Even though the report was published at the time that recombinant DNA (deoxyribonucleic acid) was becoming possible, it did not refer to this new technique and instead focused on the use and combination of existing technologies to make novel products.
Nonetheless the hitherto esoteric science of molecular biology was making considerable progress, although its practice in the early 1970s was rather distant from the world of industrial production. The phrase ‘‘genetic engineering’’ entered common parlance in the 1960s to describe human genetic modification. Medicine, however, put a premium on the use of proteins that were difficult to extract from people: insulin for diabetics and interferon for cancer sufferers. During the early 1970s what had been science fiction became fact as the use of DNA synthesis, restriction enzymes, and plasmids were integrated. In 1973 Stanley Cohen and Herbert Boyer successfully transferred a section of DNA from one E. coli bacterium to another. A few prophets such as Joshua Lederberg and Walter Gilbert argued that the new biological techniques of recombinant DNA might be ideal for making synthetic versions of expensive proteins such as insulin and interferon through their expression in bacterial cells. Small companies, such as Cetus and Genentech in California and Biogen in Cambridge, Massachusetts, were established to develop the techniques. In many cases discoveries made by small ‘‘boutique’’ companies were developed for the market by large, more established, pharmaceutical organizations.
Many governments were impressed by these advances in molecular genetics, which seemed to make biotechnology a potential counterpart to information technology in a third industrial revolution. These inspired hopes of industrial production of proteins identical to those produced in the human body that could be used to treat genetic diseases. There was also hope that industrially useful materials such as alcohol, plastics (biopolymers), or ready-colored fibers might be made in plants, and thus the attractions of a potentially new agricultural era might be as great as the implications for medicine. At a time of concern over low agricultural prices, such hopes were doubly welcome. Indeed, the agricultural benefits sometimes overshadowed the medical implications.
The mechanism for the transfer of enthusiasm from engineering fermenters to engineering genes was the New York Stock Exchange. At the end of the 1970s, new tax laws encouraged already adventurous U.S. investors to put money into small companies whose stock value might grow faster than their profits. The brokerage firm E. F. Hutton saw the potential for the new molecular biology companies such as Biogen and Cetus. Stock market interest in companies promising to make new biological entities was spurred by the 1980 decision of the U.S. Supreme Court to permit the patenting of a new organism. The patent was awarded to the Exxon researcher Ananda Chakrabarty for an organism that metabolized hydrocarbon waste. This event signaled the commercial potential of biotechnology to business and governments around the world. By the early 1980s there were widespread hopes that the protein interferon, made with some novel organism, would provide a cure for cancer. The development of monoclonal antibody technology that grew out of the work of Georges J. F. Kohler and Cesar Milstein in Cambridge (co-recipients with Niels K. Jerne of the Nobel Prize in medicine in 1986) seemed to offer new prospects for precise attacks on particular cells.
The fear of excessive regulatory controls encouraged business and scientific leaders to express optimistic projections about the potential of biotechnology. The early days of biotechnology were fired by hopes of medical products and high-value pharmaceuticals. Human insulin and interferon were early products, and a second generation included the anti-blood clotting agent tPA and the antianemia drug erythropoietin. Biotechnology was also used to help identify potential new drugs that might be made chemically, or synthetically.
At the same time agricultural products were also being developed. Three early products that each raised substantial problems were bacteria which inhibited the formation of frost on the leaves of strawberry plants (ice-minus bacteria), genetically modified plants including tomatoes and rapeseed, and the hormone bovine somatrotropin (BST) produced in genetically modified bacteria and administered to cattle in the U.S. to increase milk yields. By 1999 half the soy beans and one third of the corn grown in the U.S. were modified. Although the global spread of such products would arouse the best known concern at the end of the century, the use of the ice-minus bacteria— the first authorized release of a genetically engineered organism into the environment—had previously raised anxiety in the U.S. in the 1980s.
In 1997 Dolly the sheep was cloned from an adult mother in the Roslin agricultural research institute outside Edinburgh, Scotland. This work was inspired by the need to find a way of reproducing sheep engineered to express human proteins in their milk. However, the public interest was not so much in the cloning of sheep that had just been achieved as in the cloning of people, which had not. As in the Middle Ages when deformed creatures had been seen as monsters and portents of natural disasters, Dolly was similarly seen as monster and as a portent of human cloning.
The name Frankenstein, recalled from the story written by Mary Shelley at the beginning of the nineteenth century and from the movies of the 1930s, was once again familiar at the end of the twentieth century. Shelley had written in the shadow of Stahl’s theories. The continued appeal of this book embodies the continuity of the fears of artificial life and the anxiety over hubris. To this has been linked a more mundane suspicion of the blending of commerce and the exploitation of life. Discussion of biotechnology at the end of the twentieth century was therefore colored by questions of whose assurances of good intent and reassurance of safety could be trusted.
Browse other Technology Research Paper Topics .
ORDER HIGH QUALITY CUSTOM PAPER


Structure for writing a scientific research proposal in biotechnology

Clinical Psychology: psychopathology by systematically examining the history, classification, causes and treatment of psychological disorders.

IMPORTANCE OF LITERATURE REVIEW WRITING IN RESEARCH ARTICLE AND DISSERTATION ON BIOMEDICAL RESEARCH
- The intention of writing a research proposal in biotechnology is to get approval for research work from a committee irrespective of what you are applying.
- The researcher aims to clearly describe the research in a way that a non-specialist can understand.
- The research proposal in biotechnology must not only define how the research will be carried out but also need to provide detail description about the required timeline.
Introduction:
Writing a research proposal in the present era is an entirely challenging mission because of the constant evolution in the research design and the need to incorporate innovative concepts and medical advances in the methodology section. A well-formatted research proposal in the field of biotechnology will be written according to the required guidelines forms the mainstay for the research, and hence proposal writing is an essential step in the process of conducting research . The main objective in preparing a research proposal is to obtain approval from several committees such as the ethics committee and grant committee.
Basic requirements for an effective research proposal:
The primary purpose of every proposal that needs to address how your proposed research will fit into what is already know and the proposal should add innovative addition to the present findings in biotechnology while specifying the research question to be answered, its significance and implications. For example, TB is the deadly infectious disease in the world, but unfortunately, not all drugs are adversely affecting the Mycobacterium tuberculosis. Thus the proposing research should be innovative in finding the existing problems and identify solutions.
The research proposal must be capable of convincing the committee evaluating the proposal about the practicality, reproducibility of the proposed research in biotechnology, its achievability and credibility.
In most cases, the evaluation committee consists of the audience with different expectations and committee members from various fields such as academic professionals, policy-makers, practitioners and general audience.
In order to overcome all the challenges that come during the process of writing a quality research proposal in biotechnology, expert guidance is advisable and recommended. We have experienced professional in various fields like biotechnology , management , engineering , science and medicine , and they are ready to support in writing a full research proposal from developing innovative ideas, literature review, experimental design till budget evaluation and citation. We also provide consultations for the same and specific writing, editing , and proofreading support for your research proposal in the field of biotechnology.
See also : A Good Start For Your Research Proposal Writing
Contents of a research proposal:
In general, the research proposal format varies depending on what you are applying for, e.g. grant proposal , PhD proposal, master thesis proposal, and research position proposal. The requirements vary according to the evaluation committee and are generally provided in advance by the institution or evaluation committee.
The well-proven structure we use for writing the research proposal are:
- Aims and objectives
- Introduction
- Literature review
- Research design and methods
Title page :
Like every research document, the biotechnology research proposal will have a title page that includes.
- The title of your proposed research project (For example: “A translational recovery mechanism may provide a novel target for drug-resistant of M. tuberculosis. ”
- Name of the student
- Name of the supervisor
- The department and Institution
It is suitable for a research project, to know the specific structure for your requirements, reach out to us.
See also : Recent PhD Research Topic Ideas for Public Health
Aims and objectives :
The aim or goal and objective of the biotechnology research proposal should give a broad indication of the expected research outcome and the hypothesis to be tested can also be the aim of your study. The objective can be categorized as primary and secondary according to the parameters and tools used to achieve the goal.
Introduction :
The introduction is the initial pitch for the research proposal, and it is mandatory to explain what you want to do and why. The introduction should
- Explain the topic
- Give the background and context of the topic
- Overview of the proposed problem statement and research question
The proposal should also focus on some essential questions to guide the introduction
- Who has an interest in the topic or the target audience?
- What are the existing issues and the missing elements from the current trend?
- What new insights will your research contribute?
- Why is this research worth doing?
See also : Mastering The Art Of Writing A Cancer Biology Research Proposal
Literature review :
A literature review is the crucial part of any research work, and it helps in exploring and familiarizing much about the topic. A strong review convinces the reader and makes the audience understand the importance of the proposed research in biotechnology.
A review must demonstrate precisely how the proposed research will contribute to the technological development in biotechnology
- Compare and contrast the main theories, existing methods and controversies
- Explain the strengths and weaknesses of different approaches?
- Show how your research fits in overcoming the challenges
See also : What is a Literature review; The factors involved and its development
Research design and methods :
The objective of producing the research design and methods is to analyze how the researcher is addressing the research problem, and it is an opportunity to impress the evaluation committee by providing a well-driven research design chosen from an appropriate source. This section of the research proposal should explain the sources and methods to be incorporated in the conduct of research and discuss the specific requirements of the proposed project. In addition, the search strategy used for information gathering and analysis should be clearly explained, and it is advised to restate the main objective of the proposed project to bring back the attention as this avoids diversion. The research design and methods section should clearly describe the approach, and practical steps will be taken to address the research questions.
Research methods should state
- What tools and procedures will be used to conduct an experiment, collect and analyze data?
- Why will the chosen method be the best to answer the research questions?
Make sure not to mention a list of methods instead explain the most appropriate and reliable approach to answering the formulated research questions.
See also : Why Research Proposal Often Gets Rejected? Top 5 Reasons For Writing An Effective Research Proposal For Healthcare Dissertation
While preparing a research budget, the researcher should predict the required fund and add additional fund for unexpected delays and rising costs, and it should be justified. If the proposal is written explicitly for research funding , the researcher must categorize the budget on each part. Pubrica has experts to support in estimating the appropriate fund for any project, and we categorize each segment to determine the exact value.
Make sure to check the type of costs the funding agencies will agree to cover and include relevant items in your budget. For each category, include:
- How much money do you need?
- Why is this fund necessary to complete the research?
- How did you calculate the amount?
To determine your budget, think about:
- Materials cost
- Assistance or support staff required
- Travel costs
- Required timeline
See also : Writing a Research Grant Proposal and a Research Grant Protocol in Medical Research
Appendices:
Appendices are the support document provided for a proposal, and it will be specific for each proposal include supporting documents, questionnaires, and details of measurement tools.
Citation is the process of citing the original research paper in which the idea or methods used for composing a research proposal. And it is mandatory for any scientific research articles.
In summary, a research proposal in biotechnology should communicate the researcher’s knowledge on the project, methods and explain the need for the study. A research proposal has written for multiple reasons as mentioned, and an expert’s support will be an added advantage on any specific purpose as it gives you an edge over other participants. We provide a complete biotechnology research proposal writing service for clients who lacks background knowledge on biotechnology and specified service that you wish to hire an expert.
- K Sudheesh, Devika Rani Duggappa, and SS Nethra, How to write a research proposal? Indian J Anaesth. 2016 Sep; 60(9): 631–634.DOI: 10.4103/0019-5049.190617
- Nte AR, Awi DD, Niger, Research proposal writing: breaking the myth. J Med. 2006 Oct-Dec; 15(4):373-81.
- Labaree RV. Organizing Your Social Sciences Research Paper: Writing a Research Proposal. Available from: http://www.libguides.usc.edu/writingguide
- Guide to ALM thesis. Available from: Thesis.extension.harvard.edu/
pubrica-academy
Related posts.

Statistical analyses of case-control studies

PUB - Selecting material (e.g. excipient, active pharmaceutical ingredient) for drug development
Selecting material (e.g. excipient, active pharmaceutical ingredient, packaging material) for drug development

PUB - Health Economics of Data Modeling
Health economics in clinical trials
Comments are closed.
To revisit this article, visit My Profile, then View saved stories .
- Backchannel
- Newsletters
- WIRED Insider
- WIRED Consulting
Estelle Erasmus
How to Resist the Temptation of AI When Writing
Whether you're a student, a journalist, or a business professional, knowing how to do high-quality research and writing using trustworthy data and sources, without giving in to the temptation of AI or ChatGPT , is a skill worth developing.
As I detail in my book Writing That Gets Noticed , locating credible databases and sources and accurately vetting information can be the difference between turning a story around quickly or getting stuck with outdated information.
For example, several years ago the editor of Parents.com asked for a hot-take reaction to country singer Carrie Underwood saying that, because she was 35, she had missed her chance at having another baby. Since I had written about getting pregnant in my forties, I knew that as long as I updated my facts and figures, and included supportive and relevant peer-reviewed research, I could pull off this story. And I did.
The story ran later that day , and it led to other assignments. Here are some tips I’ve learned that you should consider mastering before you turn to automated tools like generative AI to handle your writing work for you.
Identify experts, peer-reviewed research study authors, and sources who can speak with authority—and ideally, offer easily understood sound bites or statistics on the topic of your work. Great sources include professors at major universities and media spokespeople at associations and organizations.
For example, writer and author William Dameron pinned his recent essay in HuffPost Personal around a statistic from the American Heart Association on how LGBTQ people experience higher rates of heart disease based on discrimination. Although he first found the link in a secondary source (an article in The New York Times ), he made sure that he checked the primary source: the original study that the American Heart Association gleaned the statistic from. He verified the information, as should any writer, because anytime a statistic is cited in a secondary source, errors can be introduced.
Jen Malia, author of The Infinity Rainbow Club series of children’s books (whom I recently interviewed on my podcast ), recently wrote a piece about dinosaur-bone hunting for Business Insider , which she covers in her book Violet and the Jurassic Land Exhibit.
After a visit to the Carnegie Museum of Natural History in Pittsburgh, Pennsylvania, Malia, whose books are set in Philadelphia, found multiple resources online and on the museum site that gave her the history of the Bone Wars , information on the exhibits she saw, and the scientific names of the dinosaurs she was inspired by. She also used the Library of Congress’ website, which offers digital collections and links to the Library of Congress Newspaper Collection.
Malia is a fan of searching for additional resources and citable documents with Google Scholar . “If I find that a secondary source mentions a newspaper article, I’m going to go to the original newspaper article, instead of just stopping there and quoting,” she says.
Julian Chokkattu

Chris Haslam

Boone Ashworth

Brenda Stolyar
Your local public library is a great source of free information, journals, and databases (even ones that generally require a subscription and include embargoed research). For example, your search should include everything from health databases ( Sage Journals , Scopus , PubMed) to databases for academic sources and journalism ( American Periodical Series Online , Statista , Academic Search Premier ) and databases for news, trends, market research, and polls (t he Harris Poll , Pew Research Center , Newsbank , ProPublica ).
Even if you find a study or paper that you can’t access in one of those databases, consider reaching out to the study’s lead author or researcher. In many cases, they’re happy to discuss their work and may even share the study with you directly and offer to talk about their research.
For journalist Paulette Perhach’s article on ADHD in The New York Times, she used Epic Research to see “dual team studies.” That's when two independent teams address the same topic or question, and ideally come to the same conclusions. She recommends locating research and experts via key associations for your topic. She also likes searching via Google Scholar but advises filtering it for studies and research in recent years to avoid using old data. She suggests keeping your links and research organized. “Always be ready to be peer-reviewed yourself,” Perhach says.
When you are looking for information for a story or project, you might be inclined to start with a regular Google search. But keep in mind that the internet is full of false information, and websites that look trustworthy can sometimes turn out to be businesses or companies with a vested interest in you taking their word as objective fact without additional scrutiny. Regardless of your writing project, unreliable or biased sources are a great way to torpedo your work—and any hope of future work.
Author Bobbi Rebell researched her book Launching Financial Grownups using the IRS’ website . “I might say that you can contribute a certain amount to a 401K, but it might be outdated because those numbers are always changing, and it’s important to be accurate,” she says. “AI and ChatGPT can be great for idea generation,” says Rebell, “but you have to be careful. If you are using an article someone was quoted in, you don’t know if they were misquoted or quoted out of context.”
If you use AI and ChatGPT for sourcing, you not only risk introducing errors, you risk introducing plagiarism—there is a reason OpenAI, the company behind ChatGPT, is being sued for downloading information from all those books.
Audrey Clare Farley, who writes historical nonfiction, has used a plethora of sites for historical research, including Women Also Know History , which allows searches by expertise or area of study, and JSTOR , a digital library database that offers a number of free downloads a month. She also uses Chronicling America , a project from the Library of Congress which gathers old newspapers to show how a historical event was reported, and Newspapers.com (which you can access via free trial but requires a subscription after seven days).
When it comes to finding experts, Farley cautions against choosing the loudest voices on social media platforms. “They might not necessarily be the most authoritative. I vet them by checking if they have a history of publication on the topic, and/or educational credentials.”
When vetting an expert, look for these red flags:
- You can’t find their work published or cited anywhere.
- They were published in an obscure journal.
- Their research is funded by a company, not a university, or they are the spokesperson for the company they are doing research for. (This makes them a public relations vehicle and not an appropriate source for journalism.)
And finally, the best endings for virtually any writing, whether it’s an essay, a research paper, an academic report, or a piece of investigative journalism, circle back to the beginning of the piece, and show your reader the transformation or the journey the piece has presented in perspective.
As always, your goal should be strong writing supported by research that makes an impact without cutting corners. Only then can you explore tools that might make the job a little easier, for instance by generating subheads or discovering a concept you might be missing—because then you'll have the experience and skills to see whether it's harming or helping your work.
You Might Also Like …
In your inbox: Introducing Politics Lab , your guide to election season
Google used her to tout diversity. Now she’s suing for discrimination
Our in-house physics whiz explains how heat pumps work
The big questions the Pentagon’s new UFO report fails to answer
AirPods Pro or AirPods Max? These are the best Apple buds for your ears

Michael Calore

Reece Rogers

WIRED COUPONS

Can't-Miss Deal: $100 off Dyson V12 Detect Slim Absolute cordless vacuum

GoPro Promo Code: 15% off Cameras and Accessories

Up to +30% Off with your Samsung promo code

10% Off Everything w/ Dell Coupon Code

New customers Get 25% off w/ this Vistaprint coupon

15% off Sitewide - Newegg promo code
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 26 March 2024
Predicting and improving complex beer flavor through machine learning
- Michiel Schreurs ORCID: orcid.org/0000-0002-9449-5619 1 , 2 , 3 na1 ,
- Supinya Piampongsant 1 , 2 , 3 na1 ,
- Miguel Roncoroni ORCID: orcid.org/0000-0001-7461-1427 1 , 2 , 3 na1 ,
- Lloyd Cool ORCID: orcid.org/0000-0001-9936-3124 1 , 2 , 3 , 4 ,
- Beatriz Herrera-Malaver ORCID: orcid.org/0000-0002-5096-9974 1 , 2 , 3 ,
- Christophe Vanderaa ORCID: orcid.org/0000-0001-7443-5427 4 ,
- Florian A. Theßeling 1 , 2 , 3 ,
- Łukasz Kreft ORCID: orcid.org/0000-0001-7620-4657 5 ,
- Alexander Botzki ORCID: orcid.org/0000-0001-6691-4233 5 ,
- Philippe Malcorps 6 ,
- Luk Daenen 6 ,
- Tom Wenseleers ORCID: orcid.org/0000-0002-1434-861X 4 &
- Kevin J. Verstrepen ORCID: orcid.org/0000-0002-3077-6219 1 , 2 , 3
Nature Communications volume 15 , Article number: 2368 ( 2024 ) Cite this article
42k Accesses
786 Altmetric
Metrics details
- Chemical engineering
- Gas chromatography
- Machine learning
- Metabolomics
- Taste receptors
The perception and appreciation of food flavor depends on many interacting chemical compounds and external factors, and therefore proves challenging to understand and predict. Here, we combine extensive chemical and sensory analyses of 250 different beers to train machine learning models that allow predicting flavor and consumer appreciation. For each beer, we measure over 200 chemical properties, perform quantitative descriptive sensory analysis with a trained tasting panel and map data from over 180,000 consumer reviews to train 10 different machine learning models. The best-performing algorithm, Gradient Boosting, yields models that significantly outperform predictions based on conventional statistics and accurately predict complex food features and consumer appreciation from chemical profiles. Model dissection allows identifying specific and unexpected compounds as drivers of beer flavor and appreciation. Adding these compounds results in variants of commercial alcoholic and non-alcoholic beers with improved consumer appreciation. Together, our study reveals how big data and machine learning uncover complex links between food chemistry, flavor and consumer perception, and lays the foundation to develop novel, tailored foods with superior flavors.
Similar content being viewed by others
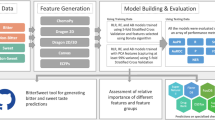
BitterSweet: Building machine learning models for predicting the bitter and sweet taste of small molecules
Rudraksh Tuwani, Somin Wadhwa & Ganesh Bagler
Sensory lexicon and aroma volatiles analysis of brewing malt
Xiaoxia Su, Miao Yu, … Tianyi Du
Predicting odor from molecular structure: a multi-label classification approach
Kushagra Saini & Venkatnarayan Ramanathan
Introduction
Predicting and understanding food perception and appreciation is one of the major challenges in food science. Accurate modeling of food flavor and appreciation could yield important opportunities for both producers and consumers, including quality control, product fingerprinting, counterfeit detection, spoilage detection, and the development of new products and product combinations (food pairing) 1 , 2 , 3 , 4 , 5 , 6 . Accurate models for flavor and consumer appreciation would contribute greatly to our scientific understanding of how humans perceive and appreciate flavor. Moreover, accurate predictive models would also facilitate and standardize existing food assessment methods and could supplement or replace assessments by trained and consumer tasting panels, which are variable, expensive and time-consuming 7 , 8 , 9 . Lastly, apart from providing objective, quantitative, accurate and contextual information that can help producers, models can also guide consumers in understanding their personal preferences 10 .
Despite the myriad of applications, predicting food flavor and appreciation from its chemical properties remains a largely elusive goal in sensory science, especially for complex food and beverages 11 , 12 . A key obstacle is the immense number of flavor-active chemicals underlying food flavor. Flavor compounds can vary widely in chemical structure and concentration, making them technically challenging and labor-intensive to quantify, even in the face of innovations in metabolomics, such as non-targeted metabolic fingerprinting 13 , 14 . Moreover, sensory analysis is perhaps even more complicated. Flavor perception is highly complex, resulting from hundreds of different molecules interacting at the physiochemical and sensorial level. Sensory perception is often non-linear, characterized by complex and concentration-dependent synergistic and antagonistic effects 15 , 16 , 17 , 18 , 19 , 20 , 21 that are further convoluted by the genetics, environment, culture and psychology of consumers 22 , 23 , 24 . Perceived flavor is therefore difficult to measure, with problems of sensitivity, accuracy, and reproducibility that can only be resolved by gathering sufficiently large datasets 25 . Trained tasting panels are considered the prime source of quality sensory data, but require meticulous training, are low throughput and high cost. Public databases containing consumer reviews of food products could provide a valuable alternative, especially for studying appreciation scores, which do not require formal training 25 . Public databases offer the advantage of amassing large amounts of data, increasing the statistical power to identify potential drivers of appreciation. However, public datasets suffer from biases, including a bias in the volunteers that contribute to the database, as well as confounding factors such as price, cult status and psychological conformity towards previous ratings of the product.
Classical multivariate statistics and machine learning methods have been used to predict flavor of specific compounds by, for example, linking structural properties of a compound to its potential biological activities or linking concentrations of specific compounds to sensory profiles 1 , 26 . Importantly, most previous studies focused on predicting organoleptic properties of single compounds (often based on their chemical structure) 27 , 28 , 29 , 30 , 31 , 32 , 33 , thus ignoring the fact that these compounds are present in a complex matrix in food or beverages and excluding complex interactions between compounds. Moreover, the classical statistics commonly used in sensory science 34 , 35 , 36 , 37 , 38 , 39 require a large sample size and sufficient variance amongst predictors to create accurate models. They are not fit for studying an extensive set of hundreds of interacting flavor compounds, since they are sensitive to outliers, have a high tendency to overfit and are less suited for non-linear and discontinuous relationships 40 .
In this study, we combine extensive chemical analyses and sensory data of a set of different commercial beers with machine learning approaches to develop models that predict taste, smell, mouthfeel and appreciation from compound concentrations. Beer is particularly suited to model the relationship between chemistry, flavor and appreciation. First, beer is a complex product, consisting of thousands of flavor compounds that partake in complex sensory interactions 41 , 42 , 43 . This chemical diversity arises from the raw materials (malt, yeast, hops, water and spices) and biochemical conversions during the brewing process (kilning, mashing, boiling, fermentation, maturation and aging) 44 , 45 . Second, the advent of the internet saw beer consumers embrace online review platforms, such as RateBeer (ZX Ventures, Anheuser-Busch InBev SA/NV) and BeerAdvocate (Next Glass, inc.). In this way, the beer community provides massive data sets of beer flavor and appreciation scores, creating extraordinarily large sensory databases to complement the analyses of our professional sensory panel. Specifically, we characterize over 200 chemical properties of 250 commercial beers, spread across 22 beer styles, and link these to the descriptive sensory profiling data of a 16-person in-house trained tasting panel and data acquired from over 180,000 public consumer reviews. These unique and extensive datasets enable us to train a suite of machine learning models to predict flavor and appreciation from a beer’s chemical profile. Dissection of the best-performing models allows us to pinpoint specific compounds as potential drivers of beer flavor and appreciation. Follow-up experiments confirm the importance of these compounds and ultimately allow us to significantly improve the flavor and appreciation of selected commercial beers. Together, our study represents a significant step towards understanding complex flavors and reinforces the value of machine learning to develop and refine complex foods. In this way, it represents a stepping stone for further computer-aided food engineering applications 46 .
To generate a comprehensive dataset on beer flavor, we selected 250 commercial Belgian beers across 22 different beer styles (Supplementary Fig. S1 ). Beers with ≤ 4.2% alcohol by volume (ABV) were classified as non-alcoholic and low-alcoholic. Blonds and Tripels constitute a significant portion of the dataset (12.4% and 11.2%, respectively) reflecting their presence on the Belgian beer market and the heterogeneity of beers within these styles. By contrast, lager beers are less diverse and dominated by a handful of brands. Rare styles such as Brut or Faro make up only a small fraction of the dataset (2% and 1%, respectively) because fewer of these beers are produced and because they are dominated by distinct characteristics in terms of flavor and chemical composition.
Extensive analysis identifies relationships between chemical compounds in beer
For each beer, we measured 226 different chemical properties, including common brewing parameters such as alcohol content, iso-alpha acids, pH, sugar concentration 47 , and over 200 flavor compounds (Methods, Supplementary Table S1 ). A large portion (37.2%) are terpenoids arising from hopping, responsible for herbal and fruity flavors 16 , 48 . A second major category are yeast metabolites, such as esters and alcohols, that result in fruity and solvent notes 48 , 49 , 50 . Other measured compounds are primarily derived from malt, or other microbes such as non- Saccharomyces yeasts and bacteria (‘wild flora’). Compounds that arise from spices or staling are labeled under ‘Others’. Five attributes (caloric value, total acids and total ester, hop aroma and sulfur compounds) are calculated from multiple individually measured compounds.
As a first step in identifying relationships between chemical properties, we determined correlations between the concentrations of the compounds (Fig. 1 , upper panel, Supplementary Data 1 and 2 , and Supplementary Fig. S2 . For the sake of clarity, only a subset of the measured compounds is shown in Fig. 1 ). Compounds of the same origin typically show a positive correlation, while absence of correlation hints at parameters varying independently. For example, the hop aroma compounds citronellol, and alpha-terpineol show moderate correlations with each other (Spearman’s rho=0.39 and 0.57), but not with the bittering hop component iso-alpha acids (Spearman’s rho=0.16 and −0.07). This illustrates how brewers can independently modify hop aroma and bitterness by selecting hop varieties and dosage time. If hops are added early in the boiling phase, chemical conversions increase bitterness while aromas evaporate, conversely, late addition of hops preserves aroma but limits bitterness 51 . Similarly, hop-derived iso-alpha acids show a strong anti-correlation with lactic acid and acetic acid, likely reflecting growth inhibition of lactic acid and acetic acid bacteria, or the consequent use of fewer hops in sour beer styles, such as West Flanders ales and Fruit beers, that rely on these bacteria for their distinct flavors 52 . Finally, yeast-derived esters (ethyl acetate, ethyl decanoate, ethyl hexanoate, ethyl octanoate) and alcohols (ethanol, isoamyl alcohol, isobutanol, and glycerol), correlate with Spearman coefficients above 0.5, suggesting that these secondary metabolites are correlated with the yeast genetic background and/or fermentation parameters and may be difficult to influence individually, although the choice of yeast strain may offer some control 53 .

Spearman rank correlations are shown. Descriptors are grouped according to their origin (malt (blue), hops (green), yeast (red), wild flora (yellow), Others (black)), and sensory aspect (aroma, taste, palate, and overall appreciation). Please note that for the chemical compounds, for the sake of clarity, only a subset of the total number of measured compounds is shown, with an emphasis on the key compounds for each source. For more details, see the main text and Methods section. Chemical data can be found in Supplementary Data 1 , correlations between all chemical compounds are depicted in Supplementary Fig. S2 and correlation values can be found in Supplementary Data 2 . See Supplementary Data 4 for sensory panel assessments and Supplementary Data 5 for correlation values between all sensory descriptors.
Interestingly, different beer styles show distinct patterns for some flavor compounds (Supplementary Fig. S3 ). These observations agree with expectations for key beer styles, and serve as a control for our measurements. For instance, Stouts generally show high values for color (darker), while hoppy beers contain elevated levels of iso-alpha acids, compounds associated with bitter hop taste. Acetic and lactic acid are not prevalent in most beers, with notable exceptions such as Kriek, Lambic, Faro, West Flanders ales and Flanders Old Brown, which use acid-producing bacteria ( Lactobacillus and Pediococcus ) or unconventional yeast ( Brettanomyces ) 54 , 55 . Glycerol, ethanol and esters show similar distributions across all beer styles, reflecting their common origin as products of yeast metabolism during fermentation 45 , 53 . Finally, low/no-alcohol beers contain low concentrations of glycerol and esters. This is in line with the production process for most of the low/no-alcohol beers in our dataset, which are produced through limiting fermentation or by stripping away alcohol via evaporation or dialysis, with both methods having the unintended side-effect of reducing the amount of flavor compounds in the final beer 56 , 57 .
Besides expected associations, our data also reveals less trivial associations between beer styles and specific parameters. For example, geraniol and citronellol, two monoterpenoids responsible for citrus, floral and rose flavors and characteristic of Citra hops, are found in relatively high amounts in Christmas, Saison, and Brett/co-fermented beers, where they may originate from terpenoid-rich spices such as coriander seeds instead of hops 58 .
Tasting panel assessments reveal sensorial relationships in beer
To assess the sensory profile of each beer, a trained tasting panel evaluated each of the 250 beers for 50 sensory attributes, including different hop, malt and yeast flavors, off-flavors and spices. Panelists used a tasting sheet (Supplementary Data 3 ) to score the different attributes. Panel consistency was evaluated by repeating 12 samples across different sessions and performing ANOVA. In 95% of cases no significant difference was found across sessions ( p > 0.05), indicating good panel consistency (Supplementary Table S2 ).
Aroma and taste perception reported by the trained panel are often linked (Fig. 1 , bottom left panel and Supplementary Data 4 and 5 ), with high correlations between hops aroma and taste (Spearman’s rho=0.83). Bitter taste was found to correlate with hop aroma and taste in general (Spearman’s rho=0.80 and 0.69), and particularly with “grassy” noble hops (Spearman’s rho=0.75). Barnyard flavor, most often associated with sour beers, is identified together with stale hops (Spearman’s rho=0.97) that are used in these beers. Lactic and acetic acid, which often co-occur, are correlated (Spearman’s rho=0.66). Interestingly, sweetness and bitterness are anti-correlated (Spearman’s rho = −0.48), confirming the hypothesis that they mask each other 59 , 60 . Beer body is highly correlated with alcohol (Spearman’s rho = 0.79), and overall appreciation is found to correlate with multiple aspects that describe beer mouthfeel (alcohol, carbonation; Spearman’s rho= 0.32, 0.39), as well as with hop and ester aroma intensity (Spearman’s rho=0.39 and 0.35).
Similar to the chemical analyses, sensorial analyses confirmed typical features of specific beer styles (Supplementary Fig. S4 ). For example, sour beers (Faro, Flanders Old Brown, Fruit beer, Kriek, Lambic, West Flanders ale) were rated acidic, with flavors of both acetic and lactic acid. Hoppy beers were found to be bitter and showed hop-associated aromas like citrus and tropical fruit. Malt taste is most detected among scotch, stout/porters, and strong ales, while low/no-alcohol beers, which often have a reputation for being ‘worty’ (reminiscent of unfermented, sweet malt extract) appear in the middle. Unsurprisingly, hop aromas are most strongly detected among hoppy beers. Like its chemical counterpart (Supplementary Fig. S3 ), acidity shows a right-skewed distribution, with the most acidic beers being Krieks, Lambics, and West Flanders ales.
Tasting panel assessments of specific flavors correlate with chemical composition
We find that the concentrations of several chemical compounds strongly correlate with specific aroma or taste, as evaluated by the tasting panel (Fig. 2 , Supplementary Fig. S5 , Supplementary Data 6 ). In some cases, these correlations confirm expectations and serve as a useful control for data quality. For example, iso-alpha acids, the bittering compounds in hops, strongly correlate with bitterness (Spearman’s rho=0.68), while ethanol and glycerol correlate with tasters’ perceptions of alcohol and body, the mouthfeel sensation of fullness (Spearman’s rho=0.82/0.62 and 0.72/0.57 respectively) and darker color from roasted malts is a good indication of malt perception (Spearman’s rho=0.54).

Heatmap colors indicate Spearman’s Rho. Axes are organized according to sensory categories (aroma, taste, mouthfeel, overall), chemical categories and chemical sources in beer (malt (blue), hops (green), yeast (red), wild flora (yellow), Others (black)). See Supplementary Data 6 for all correlation values.
Interestingly, for some relationships between chemical compounds and perceived flavor, correlations are weaker than expected. For example, the rose-smelling phenethyl acetate only weakly correlates with floral aroma. This hints at more complex relationships and interactions between compounds and suggests a need for a more complex model than simple correlations. Lastly, we uncovered unexpected correlations. For instance, the esters ethyl decanoate and ethyl octanoate appear to correlate slightly with hop perception and bitterness, possibly due to their fruity flavor. Iron is anti-correlated with hop aromas and bitterness, most likely because it is also anti-correlated with iso-alpha acids. This could be a sign of metal chelation of hop acids 61 , given that our analyses measure unbound hop acids and total iron content, or could result from the higher iron content in dark and Fruit beers, which typically have less hoppy and bitter flavors 62 .
Public consumer reviews complement expert panel data
To complement and expand the sensory data of our trained tasting panel, we collected 180,000 reviews of our 250 beers from the online consumer review platform RateBeer. This provided numerical scores for beer appearance, aroma, taste, palate, overall quality as well as the average overall score.
Public datasets are known to suffer from biases, such as price, cult status and psychological conformity towards previous ratings of a product. For example, prices correlate with appreciation scores for these online consumer reviews (rho=0.49, Supplementary Fig. S6 ), but not for our trained tasting panel (rho=0.19). This suggests that prices affect consumer appreciation, which has been reported in wine 63 , while blind tastings are unaffected. Moreover, we observe that some beer styles, like lagers and non-alcoholic beers, generally receive lower scores, reflecting that online reviewers are mostly beer aficionados with a preference for specialty beers over lager beers. In general, we find a modest correlation between our trained panel’s overall appreciation score and the online consumer appreciation scores (Fig. 3 , rho=0.29). Apart from the aforementioned biases in the online datasets, serving temperature, sample freshness and surroundings, which are all tightly controlled during the tasting panel sessions, can vary tremendously across online consumers and can further contribute to (among others, appreciation) differences between the two categories of tasters. Importantly, in contrast to the overall appreciation scores, for many sensory aspects the results from the professional panel correlated well with results obtained from RateBeer reviews. Correlations were highest for features that are relatively easy to recognize even for untrained tasters, like bitterness, sweetness, alcohol and malt aroma (Fig. 3 and below).
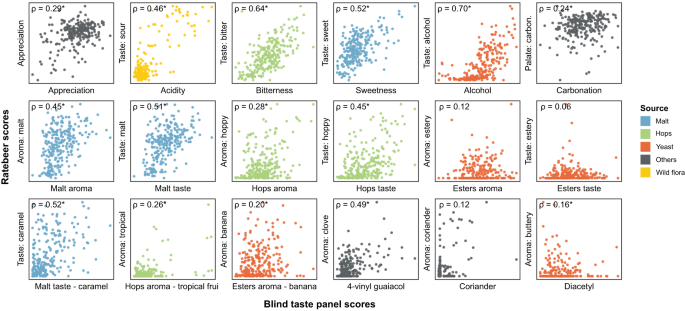
RateBeer text mining results can be found in Supplementary Data 7 . Rho values shown are Spearman correlation values, with asterisks indicating significant correlations ( p < 0.05, two-sided). All p values were smaller than 0.001, except for Esters aroma (0.0553), Esters taste (0.3275), Esters aroma—banana (0.0019), Coriander (0.0508) and Diacetyl (0.0134).
Besides collecting consumer appreciation from these online reviews, we developed automated text analysis tools to gather additional data from review texts (Supplementary Data 7 ). Processing review texts on the RateBeer database yielded comparable results to the scores given by the trained panel for many common sensory aspects, including acidity, bitterness, sweetness, alcohol, malt, and hop tastes (Fig. 3 ). This is in line with what would be expected, since these attributes require less training for accurate assessment and are less influenced by environmental factors such as temperature, serving glass and odors in the environment. Consumer reviews also correlate well with our trained panel for 4-vinyl guaiacol, a compound associated with a very characteristic aroma. By contrast, correlations for more specific aromas like ester, coriander or diacetyl are underrepresented in the online reviews, underscoring the importance of using a trained tasting panel and standardized tasting sheets with explicit factors to be scored for evaluating specific aspects of a beer. Taken together, our results suggest that public reviews are trustworthy for some, but not all, flavor features and can complement or substitute taste panel data for these sensory aspects.
Models can predict beer sensory profiles from chemical data
The rich datasets of chemical analyses, tasting panel assessments and public reviews gathered in the first part of this study provided us with a unique opportunity to develop predictive models that link chemical data to sensorial features. Given the complexity of beer flavor, basic statistical tools such as correlations or linear regression may not always be the most suitable for making accurate predictions. Instead, we applied different machine learning models that can model both simple linear and complex interactive relationships. Specifically, we constructed a set of regression models to predict (a) trained panel scores for beer flavor and quality and (b) public reviews’ appreciation scores from beer chemical profiles. We trained and tested 10 different models (Methods), 3 linear regression-based models (simple linear regression with first-order interactions (LR), lasso regression with first-order interactions (Lasso), partial least squares regressor (PLSR)), 5 decision tree models (AdaBoost regressor (ABR), extra trees (ET), gradient boosting regressor (GBR), random forest (RF) and XGBoost regressor (XGBR)), 1 support vector regression (SVR), and 1 artificial neural network (ANN) model.
To compare the performance of our machine learning models, the dataset was randomly split into a training and test set, stratified by beer style. After a model was trained on data in the training set, its performance was evaluated on its ability to predict the test dataset obtained from multi-output models (based on the coefficient of determination, see Methods). Additionally, individual-attribute models were ranked per descriptor and the average rank was calculated, as proposed by Korneva et al. 64 . Importantly, both ways of evaluating the models’ performance agreed in general. Performance of the different models varied (Table 1 ). It should be noted that all models perform better at predicting RateBeer results than results from our trained tasting panel. One reason could be that sensory data is inherently variable, and this variability is averaged out with the large number of public reviews from RateBeer. Additionally, all tree-based models perform better at predicting taste than aroma. Linear models (LR) performed particularly poorly, with negative R 2 values, due to severe overfitting (training set R 2 = 1). Overfitting is a common issue in linear models with many parameters and limited samples, especially with interaction terms further amplifying the number of parameters. L1 regularization (Lasso) successfully overcomes this overfitting, out-competing multiple tree-based models on the RateBeer dataset. Similarly, the dimensionality reduction of PLSR avoids overfitting and improves performance, to some extent. Still, tree-based models (ABR, ET, GBR, RF and XGBR) show the best performance, out-competing the linear models (LR, Lasso, PLSR) commonly used in sensory science 65 .
GBR models showed the best overall performance in predicting sensory responses from chemical information, with R 2 values up to 0.75 depending on the predicted sensory feature (Supplementary Table S4 ). The GBR models predict consumer appreciation (RateBeer) better than our trained panel’s appreciation (R 2 value of 0.67 compared to R 2 value of 0.09) (Supplementary Table S3 and Supplementary Table S4 ). ANN models showed intermediate performance, likely because neural networks typically perform best with larger datasets 66 . The SVR shows intermediate performance, mostly due to the weak predictions of specific attributes that lower the overall performance (Supplementary Table S4 ).
Model dissection identifies specific, unexpected compounds as drivers of consumer appreciation
Next, we leveraged our models to infer important contributors to sensory perception and consumer appreciation. Consumer preference is a crucial sensory aspects, because a product that shows low consumer appreciation scores often does not succeed commercially 25 . Additionally, the requirement for a large number of representative evaluators makes consumer trials one of the more costly and time-consuming aspects of product development. Hence, a model for predicting chemical drivers of overall appreciation would be a welcome addition to the available toolbox for food development and optimization.
Since GBR models on our RateBeer dataset showed the best overall performance, we focused on these models. Specifically, we used two approaches to identify important contributors. First, rankings of the most important predictors for each sensorial trait in the GBR models were obtained based on impurity-based feature importance (mean decrease in impurity). High-ranked parameters were hypothesized to be either the true causal chemical properties underlying the trait, to correlate with the actual causal properties, or to take part in sensory interactions affecting the trait 67 (Fig. 4A ). In a second approach, we used SHAP 68 to determine which parameters contributed most to the model for making predictions of consumer appreciation (Fig. 4B ). SHAP calculates parameter contributions to model predictions on a per-sample basis, which can be aggregated into an importance score.
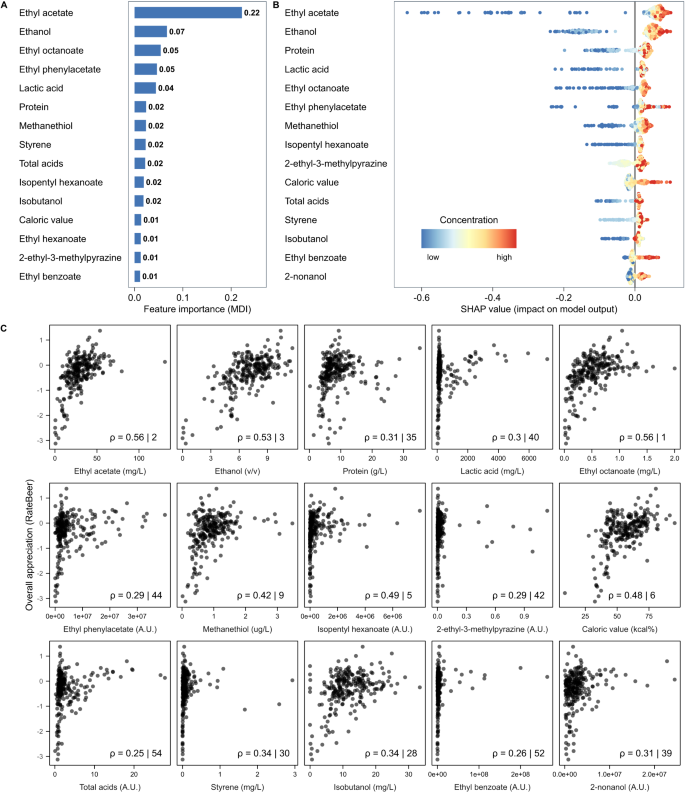
A The impurity-based feature importance (mean deviance in impurity, MDI) calculated from the Gradient Boosting Regression (GBR) model predicting RateBeer appreciation scores. The top 15 highest ranked chemical properties are shown. B SHAP summary plot for the top 15 parameters contributing to our GBR model. Each point on the graph represents a sample from our dataset. The color represents the concentration of that parameter, with bluer colors representing low values and redder colors representing higher values. Greater absolute values on the horizontal axis indicate a higher impact of the parameter on the prediction of the model. C Spearman correlations between the 15 most important chemical properties and consumer overall appreciation. Numbers indicate the Spearman Rho correlation coefficient, and the rank of this correlation compared to all other correlations. The top 15 important compounds were determined using SHAP (panel B).
Both approaches identified ethyl acetate as the most predictive parameter for beer appreciation (Fig. 4 ). Ethyl acetate is the most abundant ester in beer with a typical ‘fruity’, ‘solvent’ and ‘alcoholic’ flavor, but is often considered less important than other esters like isoamyl acetate. The second most important parameter identified by SHAP is ethanol, the most abundant beer compound after water. Apart from directly contributing to beer flavor and mouthfeel, ethanol drastically influences the physical properties of beer, dictating how easily volatile compounds escape the beer matrix to contribute to beer aroma 69 . Importantly, it should also be noted that the importance of ethanol for appreciation is likely inflated by the very low appreciation scores of non-alcoholic beers (Supplementary Fig. S4 ). Despite not often being considered a driver of beer appreciation, protein level also ranks highly in both approaches, possibly due to its effect on mouthfeel and body 70 . Lactic acid, which contributes to the tart taste of sour beers, is the fourth most important parameter identified by SHAP, possibly due to the generally high appreciation of sour beers in our dataset.
Interestingly, some of the most important predictive parameters for our model are not well-established as beer flavors or are even commonly regarded as being negative for beer quality. For example, our models identify methanethiol and ethyl phenyl acetate, an ester commonly linked to beer staling 71 , as a key factor contributing to beer appreciation. Although there is no doubt that high concentrations of these compounds are considered unpleasant, the positive effects of modest concentrations are not yet known 72 , 73 .
To compare our approach to conventional statistics, we evaluated how well the 15 most important SHAP-derived parameters correlate with consumer appreciation (Fig. 4C ). Interestingly, only 6 of the properties derived by SHAP rank amongst the top 15 most correlated parameters. For some chemical compounds, the correlations are so low that they would have likely been considered unimportant. For example, lactic acid, the fourth most important parameter, shows a bimodal distribution for appreciation, with sour beers forming a separate cluster, that is missed entirely by the Spearman correlation. Additionally, the correlation plots reveal outliers, emphasizing the need for robust analysis tools. Together, this highlights the need for alternative models, like the Gradient Boosting model, that better grasp the complexity of (beer) flavor.
Finally, to observe the relationships between these chemical properties and their predicted targets, partial dependence plots were constructed for the six most important predictors of consumer appreciation 74 , 75 , 76 (Supplementary Fig. S7 ). One-way partial dependence plots show how a change in concentration affects the predicted appreciation. These plots reveal an important limitation of our models: appreciation predictions remain constant at ever-increasing concentrations. This implies that once a threshold concentration is reached, further increasing the concentration does not affect appreciation. This is false, as it is well-documented that certain compounds become unpleasant at high concentrations, including ethyl acetate (‘nail polish’) 77 and methanethiol (‘sulfury’ and ‘rotten cabbage’) 78 . The inability of our models to grasp that flavor compounds have optimal levels, above which they become negative, is a consequence of working with commercial beer brands where (off-)flavors are rarely too high to negatively impact the product. The two-way partial dependence plots show how changing the concentration of two compounds influences predicted appreciation, visualizing their interactions (Supplementary Fig. S7 ). In our case, the top 5 parameters are dominated by additive or synergistic interactions, with high concentrations for both compounds resulting in the highest predicted appreciation.
To assess the robustness of our best-performing models and model predictions, we performed 100 iterations of the GBR, RF and ET models. In general, all iterations of the models yielded similar performance (Supplementary Fig. S8 ). Moreover, the main predictors (including the top predictors ethanol and ethyl acetate) remained virtually the same, especially for GBR and RF. For the iterations of the ET model, we did observe more variation in the top predictors, which is likely a consequence of the model’s inherent random architecture in combination with co-correlations between certain predictors. However, even in this case, several of the top predictors (ethanol and ethyl acetate) remain unchanged, although their rank in importance changes (Supplementary Fig. S8 ).
Next, we investigated if a combination of RateBeer and trained panel data into one consolidated dataset would lead to stronger models, under the hypothesis that such a model would suffer less from bias in the datasets. A GBR model was trained to predict appreciation on the combined dataset. This model underperformed compared to the RateBeer model, both in the native case and when including a dataset identifier (R 2 = 0.67, 0.26 and 0.42 respectively). For the latter, the dataset identifier is the most important feature (Supplementary Fig. S9 ), while most of the feature importance remains unchanged, with ethyl acetate and ethanol ranking highest, like in the original model trained only on RateBeer data. It seems that the large variation in the panel dataset introduces noise, weakening the models’ performances and reliability. In addition, it seems reasonable to assume that both datasets are fundamentally different, with the panel dataset obtained by blind tastings by a trained professional panel.
Lastly, we evaluated whether beer style identifiers would further enhance the model’s performance. A GBR model was trained with parameters that explicitly encoded the styles of the samples. This did not improve model performance (R2 = 0.66 with style information vs R2 = 0.67). The most important chemical features are consistent with the model trained without style information (eg. ethanol and ethyl acetate), and with the exception of the most preferred (strong ale) and least preferred (low/no-alcohol) styles, none of the styles were among the most important features (Supplementary Fig. S9 , Supplementary Table S5 and S6 ). This is likely due to a combination of style-specific chemical signatures, such as iso-alpha acids and lactic acid, that implicitly convey style information to the original models, as well as the low number of samples belonging to some styles, making it difficult for the model to learn style-specific patterns. Moreover, beer styles are not rigorously defined, with some styles overlapping in features and some beers being misattributed to a specific style, all of which leads to more noise in models that use style parameters.
Model validation
To test if our predictive models give insight into beer appreciation, we set up experiments aimed at improving existing commercial beers. We specifically selected overall appreciation as the trait to be examined because of its complexity and commercial relevance. Beer flavor comprises a complex bouquet rather than single aromas and tastes 53 . Hence, adding a single compound to the extent that a difference is noticeable may lead to an unbalanced, artificial flavor. Therefore, we evaluated the effect of combinations of compounds. Because Blond beers represent the most extensive style in our dataset, we selected a beer from this style as the starting material for these experiments (Beer 64 in Supplementary Data 1 ).
In the first set of experiments, we adjusted the concentrations of compounds that made up the most important predictors of overall appreciation (ethyl acetate, ethanol, lactic acid, ethyl phenyl acetate) together with correlated compounds (ethyl hexanoate, isoamyl acetate, glycerol), bringing them up to 95 th percentile ethanol-normalized concentrations (Methods) within the Blond group (‘Spiked’ concentration in Fig. 5A ). Compared to controls, the spiked beers were found to have significantly improved overall appreciation among trained panelists, with panelist noting increased intensity of ester flavors, sweetness, alcohol, and body fullness (Fig. 5B ). To disentangle the contribution of ethanol to these results, a second experiment was performed without the addition of ethanol. This resulted in a similar outcome, including increased perception of alcohol and overall appreciation.
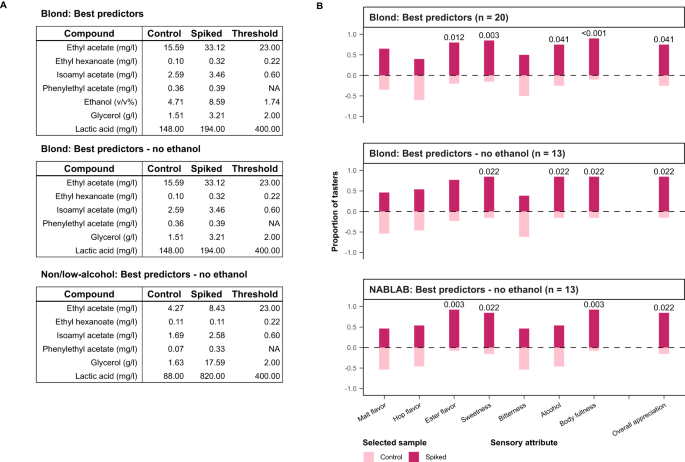
Adding the top chemical compounds, identified as best predictors of appreciation by our model, into poorly appreciated beers results in increased appreciation from our trained panel. Results of sensory tests between base beers and those spiked with compounds identified as the best predictors by the model. A Blond and Non/Low-alcohol (0.0% ABV) base beers were brought up to 95th-percentile ethanol-normalized concentrations within each style. B For each sensory attribute, tasters indicated the more intense sample and selected the sample they preferred. The numbers above the bars correspond to the p values that indicate significant changes in perceived flavor (two-sided binomial test: alpha 0.05, n = 20 or 13).
In a last experiment, we tested whether using the model’s predictions can boost the appreciation of a non-alcoholic beer (beer 223 in Supplementary Data 1 ). Again, the addition of a mixture of predicted compounds (omitting ethanol, in this case) resulted in a significant increase in appreciation, body, ester flavor and sweetness.
Predicting flavor and consumer appreciation from chemical composition is one of the ultimate goals of sensory science. A reliable, systematic and unbiased way to link chemical profiles to flavor and food appreciation would be a significant asset to the food and beverage industry. Such tools would substantially aid in quality control and recipe development, offer an efficient and cost-effective alternative to pilot studies and consumer trials and would ultimately allow food manufacturers to produce superior, tailor-made products that better meet the demands of specific consumer groups more efficiently.
A limited set of studies have previously tried, to varying degrees of success, to predict beer flavor and beer popularity based on (a limited set of) chemical compounds and flavors 79 , 80 . Current sensitive, high-throughput technologies allow measuring an unprecedented number of chemical compounds and properties in a large set of samples, yielding a dataset that can train models that help close the gaps between chemistry and flavor, even for a complex natural product like beer. To our knowledge, no previous research gathered data at this scale (250 samples, 226 chemical parameters, 50 sensory attributes and 5 consumer scores) to disentangle and validate the chemical aspects driving beer preference using various machine-learning techniques. We find that modern machine learning models outperform conventional statistical tools, such as correlations and linear models, and can successfully predict flavor appreciation from chemical composition. This could be attributed to the natural incorporation of interactions and non-linear or discontinuous effects in machine learning models, which are not easily grasped by the linear model architecture. While linear models and partial least squares regression represent the most widespread statistical approaches in sensory science, in part because they allow interpretation 65 , 81 , 82 , modern machine learning methods allow for building better predictive models while preserving the possibility to dissect and exploit the underlying patterns. Of the 10 different models we trained, tree-based models, such as our best performing GBR, showed the best overall performance in predicting sensory responses from chemical information, outcompeting artificial neural networks. This agrees with previous reports for models trained on tabular data 83 . Our results are in line with the findings of Colantonio et al. who also identified the gradient boosting architecture as performing best at predicting appreciation and flavor (of tomatoes and blueberries, in their specific study) 26 . Importantly, besides our larger experimental scale, we were able to directly confirm our models’ predictions in vivo.
Our study confirms that flavor compound concentration does not always correlate with perception, suggesting complex interactions that are often missed by more conventional statistics and simple models. Specifically, we find that tree-based algorithms may perform best in developing models that link complex food chemistry with aroma. Furthermore, we show that massive datasets of untrained consumer reviews provide a valuable source of data, that can complement or even replace trained tasting panels, especially for appreciation and basic flavors, such as sweetness and bitterness. This holds despite biases that are known to occur in such datasets, such as price or conformity bias. Moreover, GBR models predict taste better than aroma. This is likely because taste (e.g. bitterness) often directly relates to the corresponding chemical measurements (e.g., iso-alpha acids), whereas such a link is less clear for aromas, which often result from the interplay between multiple volatile compounds. We also find that our models are best at predicting acidity and alcohol, likely because there is a direct relation between the measured chemical compounds (acids and ethanol) and the corresponding perceived sensorial attribute (acidity and alcohol), and because even untrained consumers are generally able to recognize these flavors and aromas.
The predictions of our final models, trained on review data, hold even for blind tastings with small groups of trained tasters, as demonstrated by our ability to validate specific compounds as drivers of beer flavor and appreciation. Since adding a single compound to the extent of a noticeable difference may result in an unbalanced flavor profile, we specifically tested our identified key drivers as a combination of compounds. While this approach does not allow us to validate if a particular single compound would affect flavor and/or appreciation, our experiments do show that this combination of compounds increases consumer appreciation.
It is important to stress that, while it represents an important step forward, our approach still has several major limitations. A key weakness of the GBR model architecture is that amongst co-correlating variables, the largest main effect is consistently preferred for model building. As a result, co-correlating variables often have artificially low importance scores, both for impurity and SHAP-based methods, like we observed in the comparison to the more randomized Extra Trees models. This implies that chemicals identified as key drivers of a specific sensory feature by GBR might not be the true causative compounds, but rather co-correlate with the actual causative chemical. For example, the high importance of ethyl acetate could be (partially) attributed to the total ester content, ethanol or ethyl hexanoate (rho=0.77, rho=0.72 and rho=0.68), while ethyl phenylacetate could hide the importance of prenyl isobutyrate and ethyl benzoate (rho=0.77 and rho=0.76). Expanding our GBR model to include beer style as a parameter did not yield additional power or insight. This is likely due to style-specific chemical signatures, such as iso-alpha acids and lactic acid, that implicitly convey style information to the original model, as well as the smaller sample size per style, limiting the power to uncover style-specific patterns. This can be partly attributed to the curse of dimensionality, where the high number of parameters results in the models mainly incorporating single parameter effects, rather than complex interactions such as style-dependent effects 67 . A larger number of samples may overcome some of these limitations and offer more insight into style-specific effects. On the other hand, beer style is not a rigid scientific classification, and beers within one style often differ a lot, which further complicates the analysis of style as a model factor.
Our study is limited to beers from Belgian breweries. Although these beers cover a large portion of the beer styles available globally, some beer styles and consumer patterns may be missing, while other features might be overrepresented. For example, many Belgian ales exhibit yeast-driven flavor profiles, which is reflected in the chemical drivers of appreciation discovered by this study. In future work, expanding the scope to include diverse markets and beer styles could lead to the identification of even more drivers of appreciation and better models for special niche products that were not present in our beer set.
In addition to inherent limitations of GBR models, there are also some limitations associated with studying food aroma. Even if our chemical analyses measured most of the known aroma compounds, the total number of flavor compounds in complex foods like beer is still larger than the subset we were able to measure in this study. For example, hop-derived thiols, that influence flavor at very low concentrations, are notoriously difficult to measure in a high-throughput experiment. Moreover, consumer perception remains subjective and prone to biases that are difficult to avoid. It is also important to stress that the models are still immature and that more extensive datasets will be crucial for developing more complete models in the future. Besides more samples and parameters, our dataset does not include any demographic information about the tasters. Including such data could lead to better models that grasp external factors like age and culture. Another limitation is that our set of beers consists of high-quality end-products and lacks beers that are unfit for sale, which limits the current model in accurately predicting products that are appreciated very badly. Finally, while models could be readily applied in quality control, their use in sensory science and product development is restrained by their inability to discern causal relationships. Given that the models cannot distinguish compounds that genuinely drive consumer perception from those that merely correlate, validation experiments are essential to identify true causative compounds.
Despite the inherent limitations, dissection of our models enabled us to pinpoint specific molecules as potential drivers of beer aroma and consumer appreciation, including compounds that were unexpected and would not have been identified using standard approaches. Important drivers of beer appreciation uncovered by our models include protein levels, ethyl acetate, ethyl phenyl acetate and lactic acid. Currently, many brewers already use lactic acid to acidify their brewing water and ensure optimal pH for enzymatic activity during the mashing process. Our results suggest that adding lactic acid can also improve beer appreciation, although its individual effect remains to be tested. Interestingly, ethanol appears to be unnecessary to improve beer appreciation, both for blond beer and alcohol-free beer. Given the growing consumer interest in alcohol-free beer, with a predicted annual market growth of >7% 84 , it is relevant for brewers to know what compounds can further increase consumer appreciation of these beers. Hence, our model may readily provide avenues to further improve the flavor and consumer appreciation of both alcoholic and non-alcoholic beers, which is generally considered one of the key challenges for future beer production.
Whereas we see a direct implementation of our results for the development of superior alcohol-free beverages and other food products, our study can also serve as a stepping stone for the development of novel alcohol-containing beverages. We want to echo the growing body of scientific evidence for the negative effects of alcohol consumption, both on the individual level by the mutagenic, teratogenic and carcinogenic effects of ethanol 85 , 86 , as well as the burden on society caused by alcohol abuse and addiction. We encourage the use of our results for the production of healthier, tastier products, including novel and improved beverages with lower alcohol contents. Furthermore, we strongly discourage the use of these technologies to improve the appreciation or addictive properties of harmful substances.
The present work demonstrates that despite some important remaining hurdles, combining the latest developments in chemical analyses, sensory analysis and modern machine learning methods offers exciting avenues for food chemistry and engineering. Soon, these tools may provide solutions in quality control and recipe development, as well as new approaches to sensory science and flavor research.
Beer selection
250 commercial Belgian beers were selected to cover the broad diversity of beer styles and corresponding diversity in chemical composition and aroma. See Supplementary Fig. S1 .
Chemical dataset
Sample preparation.
Beers within their expiration date were purchased from commercial retailers. Samples were prepared in biological duplicates at room temperature, unless explicitly stated otherwise. Bottle pressure was measured with a manual pressure device (Steinfurth Mess-Systeme GmbH) and used to calculate CO 2 concentration. The beer was poured through two filter papers (Macherey-Nagel, 500713032 MN 713 ¼) to remove carbon dioxide and prevent spontaneous foaming. Samples were then prepared for measurements by targeted Headspace-Gas Chromatography-Flame Ionization Detector/Flame Photometric Detector (HS-GC-FID/FPD), Headspace-Solid Phase Microextraction-Gas Chromatography-Mass Spectrometry (HS-SPME-GC-MS), colorimetric analysis, enzymatic analysis, Near-Infrared (NIR) analysis, as described in the sections below. The mean values of biological duplicates are reported for each compound.
HS-GC-FID/FPD
HS-GC-FID/FPD (Shimadzu GC 2010 Plus) was used to measure higher alcohols, acetaldehyde, esters, 4-vinyl guaicol, and sulfur compounds. Each measurement comprised 5 ml of sample pipetted into a 20 ml glass vial containing 1.75 g NaCl (VWR, 27810.295). 100 µl of 2-heptanol (Sigma-Aldrich, H3003) (internal standard) solution in ethanol (Fisher Chemical, E/0650DF/C17) was added for a final concentration of 2.44 mg/L. Samples were flushed with nitrogen for 10 s, sealed with a silicone septum, stored at −80 °C and analyzed in batches of 20.
The GC was equipped with a DB-WAXetr column (length, 30 m; internal diameter, 0.32 mm; layer thickness, 0.50 µm; Agilent Technologies, Santa Clara, CA, USA) to the FID and an HP-5 column (length, 30 m; internal diameter, 0.25 mm; layer thickness, 0.25 µm; Agilent Technologies, Santa Clara, CA, USA) to the FPD. N 2 was used as the carrier gas. Samples were incubated for 20 min at 70 °C in the headspace autosampler (Flow rate, 35 cm/s; Injection volume, 1000 µL; Injection mode, split; Combi PAL autosampler, CTC analytics, Switzerland). The injector, FID and FPD temperatures were kept at 250 °C. The GC oven temperature was first held at 50 °C for 5 min and then allowed to rise to 80 °C at a rate of 5 °C/min, followed by a second ramp of 4 °C/min until 200 °C kept for 3 min and a final ramp of (4 °C/min) until 230 °C for 1 min. Results were analyzed with the GCSolution software version 2.4 (Shimadzu, Kyoto, Japan). The GC was calibrated with a 5% EtOH solution (VWR International) containing the volatiles under study (Supplementary Table S7 ).
HS-SPME-GC-MS
HS-SPME-GC-MS (Shimadzu GCMS-QP-2010 Ultra) was used to measure additional volatile compounds, mainly comprising terpenoids and esters. Samples were analyzed by HS-SPME using a triphase DVB/Carboxen/PDMS 50/30 μm SPME fiber (Supelco Co., Bellefonte, PA, USA) followed by gas chromatography (Thermo Fisher Scientific Trace 1300 series, USA) coupled to a mass spectrometer (Thermo Fisher Scientific ISQ series MS) equipped with a TriPlus RSH autosampler. 5 ml of degassed beer sample was placed in 20 ml vials containing 1.75 g NaCl (VWR, 27810.295). 5 µl internal standard mix was added, containing 2-heptanol (1 g/L) (Sigma-Aldrich, H3003), 4-fluorobenzaldehyde (1 g/L) (Sigma-Aldrich, 128376), 2,3-hexanedione (1 g/L) (Sigma-Aldrich, 144169) and guaiacol (1 g/L) (Sigma-Aldrich, W253200) in ethanol (Fisher Chemical, E/0650DF/C17). Each sample was incubated at 60 °C in the autosampler oven with constant agitation. After 5 min equilibration, the SPME fiber was exposed to the sample headspace for 30 min. The compounds trapped on the fiber were thermally desorbed in the injection port of the chromatograph by heating the fiber for 15 min at 270 °C.
The GC-MS was equipped with a low polarity RXi-5Sil MS column (length, 20 m; internal diameter, 0.18 mm; layer thickness, 0.18 µm; Restek, Bellefonte, PA, USA). Injection was performed in splitless mode at 320 °C, a split flow of 9 ml/min, a purge flow of 5 ml/min and an open valve time of 3 min. To obtain a pulsed injection, a programmed gas flow was used whereby the helium gas flow was set at 2.7 mL/min for 0.1 min, followed by a decrease in flow of 20 ml/min to the normal 0.9 mL/min. The temperature was first held at 30 °C for 3 min and then allowed to rise to 80 °C at a rate of 7 °C/min, followed by a second ramp of 2 °C/min till 125 °C and a final ramp of 8 °C/min with a final temperature of 270 °C.
Mass acquisition range was 33 to 550 amu at a scan rate of 5 scans/s. Electron impact ionization energy was 70 eV. The interface and ion source were kept at 275 °C and 250 °C, respectively. A mix of linear n-alkanes (from C7 to C40, Supelco Co.) was injected into the GC-MS under identical conditions to serve as external retention index markers. Identification and quantification of the compounds were performed using an in-house developed R script as described in Goelen et al. and Reher et al. 87 , 88 (for package information, see Supplementary Table S8 ). Briefly, chromatograms were analyzed using AMDIS (v2.71) 89 to separate overlapping peaks and obtain pure compound spectra. The NIST MS Search software (v2.0 g) in combination with the NIST2017, FFNSC3 and Adams4 libraries were used to manually identify the empirical spectra, taking into account the expected retention time. After background subtraction and correcting for retention time shifts between samples run on different days based on alkane ladders, compound elution profiles were extracted and integrated using a file with 284 target compounds of interest, which were either recovered in our identified AMDIS list of spectra or were known to occur in beer. Compound elution profiles were estimated for every peak in every chromatogram over a time-restricted window using weighted non-negative least square analysis after which peak areas were integrated 87 , 88 . Batch effect correction was performed by normalizing against the most stable internal standard compound, 4-fluorobenzaldehyde. Out of all 284 target compounds that were analyzed, 167 were visually judged to have reliable elution profiles and were used for final analysis.
Discrete photometric and enzymatic analysis
Discrete photometric and enzymatic analysis (Thermo Scientific TM Gallery TM Plus Beermaster Discrete Analyzer) was used to measure acetic acid, ammonia, beta-glucan, iso-alpha acids, color, sugars, glycerol, iron, pH, protein, and sulfite. 2 ml of sample volume was used for the analyses. Information regarding the reagents and standard solutions used for analyses and calibrations is included in Supplementary Table S7 and Supplementary Table S9 .
NIR analyses
NIR analysis (Anton Paar Alcolyzer Beer ME System) was used to measure ethanol. Measurements comprised 50 ml of sample, and a 10% EtOH solution was used for calibration.
Correlation calculations
Pairwise Spearman Rank correlations were calculated between all chemical properties.
Sensory dataset
Trained panel.
Our trained tasting panel consisted of volunteers who gave prior verbal informed consent. All compounds used for the validation experiment were of food-grade quality. The tasting sessions were approved by the Social and Societal Ethics Committee of the KU Leuven (G-2022-5677-R2(MAR)). All online reviewers agreed to the Terms and Conditions of the RateBeer website.
Sensory analysis was performed according to the American Society of Brewing Chemists (ASBC) Sensory Analysis Methods 90 . 30 volunteers were screened through a series of triangle tests. The sixteen most sensitive and consistent tasters were retained as taste panel members. The resulting panel was diverse in age [22–42, mean: 29], sex [56% male] and nationality [7 different countries]. The panel developed a consensus vocabulary to describe beer aroma, taste and mouthfeel. Panelists were trained to identify and score 50 different attributes, using a 7-point scale to rate attributes’ intensity. The scoring sheet is included as Supplementary Data 3 . Sensory assessments took place between 10–12 a.m. The beers were served in black-colored glasses. Per session, between 5 and 12 beers of the same style were tasted at 12 °C to 16 °C. Two reference beers were added to each set and indicated as ‘Reference 1 & 2’, allowing panel members to calibrate their ratings. Not all panelists were present at every tasting. Scores were scaled by standard deviation and mean-centered per taster. Values are represented as z-scores and clustered by Euclidean distance. Pairwise Spearman correlations were calculated between taste and aroma sensory attributes. Panel consistency was evaluated by repeating samples on different sessions and performing ANOVA to identify differences, using the ‘stats’ package (v4.2.2) in R (for package information, see Supplementary Table S8 ).
Online reviews from a public database
The ‘scrapy’ package in Python (v3.6) (for package information, see Supplementary Table S8 ). was used to collect 232,288 online reviews (mean=922, min=6, max=5343) from RateBeer, an online beer review database. Each review entry comprised 5 numerical scores (appearance, aroma, taste, palate and overall quality) and an optional review text. The total number of reviews per reviewer was collected separately. Numerical scores were scaled and centered per rater, and mean scores were calculated per beer.
For the review texts, the language was estimated using the packages ‘langdetect’ and ‘langid’ in Python. Reviews that were classified as English by both packages were kept. Reviewers with fewer than 100 entries overall were discarded. 181,025 reviews from >6000 reviewers from >40 countries remained. Text processing was done using the ‘nltk’ package in Python. Texts were corrected for slang and misspellings; proper nouns and rare words that are relevant to the beer context were specified and kept as-is (‘Chimay’,’Lambic’, etc.). A dictionary of semantically similar sensorial terms, for example ‘floral’ and ‘flower’, was created and collapsed together into one term. Words were stemmed and lemmatized to avoid identifying words such as ‘acid’ and ‘acidity’ as separate terms. Numbers and punctuation were removed.
Sentences from up to 50 randomly chosen reviews per beer were manually categorized according to the aspect of beer they describe (appearance, aroma, taste, palate, overall quality—not to be confused with the 5 numerical scores described above) or flagged as irrelevant if they contained no useful information. If a beer contained fewer than 50 reviews, all reviews were manually classified. This labeled data set was used to train a model that classified the rest of the sentences for all beers 91 . Sentences describing taste and aroma were extracted, and term frequency–inverse document frequency (TFIDF) was implemented to calculate enrichment scores for sensorial words per beer.
The sex of the tasting subject was not considered when building our sensory database. Instead, results from different panelists were averaged, both for our trained panel (56% male, 44% female) and the RateBeer reviews (70% male, 30% female for RateBeer as a whole).
Beer price collection and processing
Beer prices were collected from the following stores: Colruyt, Delhaize, Total Wine, BeerHawk, The Belgian Beer Shop, The Belgian Shop, and Beer of Belgium. Where applicable, prices were converted to Euros and normalized per liter. Spearman correlations were calculated between these prices and mean overall appreciation scores from RateBeer and the taste panel, respectively.
Pairwise Spearman Rank correlations were calculated between all sensory properties.
Machine learning models
Predictive modeling of sensory profiles from chemical data.
Regression models were constructed to predict (a) trained panel scores for beer flavors and quality from beer chemical profiles and (b) public reviews’ appreciation scores from beer chemical profiles. Z-scores were used to represent sensory attributes in both data sets. Chemical properties with log-normal distributions (Shapiro-Wilk test, p < 0.05 ) were log-transformed. Missing chemical measurements (0.1% of all data) were replaced with mean values per attribute. Observations from 250 beers were randomly separated into a training set (70%, 175 beers) and a test set (30%, 75 beers), stratified per beer style. Chemical measurements (p = 231) were normalized based on the training set average and standard deviation. In total, three linear regression-based models: linear regression with first-order interaction terms (LR), lasso regression with first-order interaction terms (Lasso) and partial least squares regression (PLSR); five decision tree models, Adaboost regressor (ABR), Extra Trees (ET), Gradient Boosting regressor (GBR), Random Forest (RF) and XGBoost regressor (XGBR); one support vector machine model (SVR) and one artificial neural network model (ANN) were trained. The models were implemented using the ‘scikit-learn’ package (v1.2.2) and ‘xgboost’ package (v1.7.3) in Python (v3.9.16). Models were trained, and hyperparameters optimized, using five-fold cross-validated grid search with the coefficient of determination (R 2 ) as the evaluation metric. The ANN (scikit-learn’s MLPRegressor) was optimized using Bayesian Tree-Structured Parzen Estimator optimization with the ‘Optuna’ Python package (v3.2.0). Individual models were trained per attribute, and a multi-output model was trained on all attributes simultaneously.
Model dissection
GBR was found to outperform other methods, resulting in models with the highest average R 2 values in both trained panel and public review data sets. Impurity-based rankings of the most important predictors for each predicted sensorial trait were obtained using the ‘scikit-learn’ package. To observe the relationships between these chemical properties and their predicted targets, partial dependence plots (PDP) were constructed for the six most important predictors of consumer appreciation 74 , 75 .
The ‘SHAP’ package in Python (v0.41.0) was implemented to provide an alternative ranking of predictor importance and to visualize the predictors’ effects as a function of their concentration 68 .
Validation of causal chemical properties
To validate the effects of the most important model features on predicted sensory attributes, beers were spiked with the chemical compounds identified by the models and descriptive sensory analyses were carried out according to the American Society of Brewing Chemists (ASBC) protocol 90 .
Compound spiking was done 30 min before tasting. Compounds were spiked into fresh beer bottles, that were immediately resealed and inverted three times. Fresh bottles of beer were opened for the same duration, resealed, and inverted thrice, to serve as controls. Pairs of spiked samples and controls were served simultaneously, chilled and in dark glasses as outlined in the Trained panel section above. Tasters were instructed to select the glass with the higher flavor intensity for each attribute (directional difference test 92 ) and to select the glass they prefer.
The final concentration after spiking was equal to the within-style average, after normalizing by ethanol concentration. This was done to ensure balanced flavor profiles in the final spiked beer. The same methods were applied to improve a non-alcoholic beer. Compounds were the following: ethyl acetate (Merck KGaA, W241415), ethyl hexanoate (Merck KGaA, W243906), isoamyl acetate (Merck KGaA, W205508), phenethyl acetate (Merck KGaA, W285706), ethanol (96%, Colruyt), glycerol (Merck KGaA, W252506), lactic acid (Merck KGaA, 261106).
Significant differences in preference or perceived intensity were determined by performing the two-sided binomial test on each attribute.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support the findings of this work are available in the Supplementary Data files and have been deposited to Zenodo under accession code 10653704 93 . The RateBeer scores data are under restricted access, they are not publicly available as they are property of RateBeer (ZX Ventures, USA). Access can be obtained from the authors upon reasonable request and with permission of RateBeer (ZX Ventures, USA). Source data are provided with this paper.
Code availability
The code for training the machine learning models, analyzing the models, and generating the figures has been deposited to Zenodo under accession code 10653704 93 .
Tieman, D. et al. A chemical genetic roadmap to improved tomato flavor. Science 355 , 391–394 (2017).
Article ADS CAS PubMed Google Scholar
Plutowska, B. & Wardencki, W. Application of gas chromatography–olfactometry (GC–O) in analysis and quality assessment of alcoholic beverages – A review. Food Chem. 107 , 449–463 (2008).
Article CAS Google Scholar
Legin, A., Rudnitskaya, A., Seleznev, B. & Vlasov, Y. Electronic tongue for quality assessment of ethanol, vodka and eau-de-vie. Anal. Chim. Acta 534 , 129–135 (2005).
Loutfi, A., Coradeschi, S., Mani, G. K., Shankar, P. & Rayappan, J. B. B. Electronic noses for food quality: A review. J. Food Eng. 144 , 103–111 (2015).
Ahn, Y.-Y., Ahnert, S. E., Bagrow, J. P. & Barabási, A.-L. Flavor network and the principles of food pairing. Sci. Rep. 1 , 196 (2011).
Article CAS PubMed PubMed Central Google Scholar
Bartoshuk, L. M. & Klee, H. J. Better fruits and vegetables through sensory analysis. Curr. Biol. 23 , R374–R378 (2013).
Article CAS PubMed Google Scholar
Piggott, J. R. Design questions in sensory and consumer science. Food Qual. Prefer. 3293 , 217–220 (1995).
Article Google Scholar
Kermit, M. & Lengard, V. Assessing the performance of a sensory panel-panellist monitoring and tracking. J. Chemom. 19 , 154–161 (2005).
Cook, D. J., Hollowood, T. A., Linforth, R. S. T. & Taylor, A. J. Correlating instrumental measurements of texture and flavour release with human perception. Int. J. Food Sci. Technol. 40 , 631–641 (2005).
Chinchanachokchai, S., Thontirawong, P. & Chinchanachokchai, P. A tale of two recommender systems: The moderating role of consumer expertise on artificial intelligence based product recommendations. J. Retail. Consum. Serv. 61 , 1–12 (2021).
Ross, C. F. Sensory science at the human-machine interface. Trends Food Sci. Technol. 20 , 63–72 (2009).
Chambers, E. IV & Koppel, K. Associations of volatile compounds with sensory aroma and flavor: The complex nature of flavor. Molecules 18 , 4887–4905 (2013).
Pinu, F. R. Metabolomics—The new frontier in food safety and quality research. Food Res. Int. 72 , 80–81 (2015).
Danezis, G. P., Tsagkaris, A. S., Brusic, V. & Georgiou, C. A. Food authentication: state of the art and prospects. Curr. Opin. Food Sci. 10 , 22–31 (2016).
Shepherd, G. M. Smell images and the flavour system in the human brain. Nature 444 , 316–321 (2006).
Meilgaard, M. C. Prediction of flavor differences between beers from their chemical composition. J. Agric. Food Chem. 30 , 1009–1017 (1982).
Xu, L. et al. Widespread receptor-driven modulation in peripheral olfactory coding. Science 368 , eaaz5390 (2020).
Kupferschmidt, K. Following the flavor. Science 340 , 808–809 (2013).
Billesbølle, C. B. et al. Structural basis of odorant recognition by a human odorant receptor. Nature 615 , 742–749 (2023).
Article ADS PubMed PubMed Central Google Scholar
Smith, B. Perspective: Complexities of flavour. Nature 486 , S6–S6 (2012).
Pfister, P. et al. Odorant receptor inhibition is fundamental to odor encoding. Curr. Biol. 30 , 2574–2587 (2020).
Moskowitz, H. W., Kumaraiah, V., Sharma, K. N., Jacobs, H. L. & Sharma, S. D. Cross-cultural differences in simple taste preferences. Science 190 , 1217–1218 (1975).
Eriksson, N. et al. A genetic variant near olfactory receptor genes influences cilantro preference. Flavour 1 , 22 (2012).
Ferdenzi, C. et al. Variability of affective responses to odors: Culture, gender, and olfactory knowledge. Chem. Senses 38 , 175–186 (2013).
Article PubMed Google Scholar
Lawless, H. T. & Heymann, H. Sensory evaluation of food: Principles and practices. (Springer, New York, NY). https://doi.org/10.1007/978-1-4419-6488-5 (2010).
Colantonio, V. et al. Metabolomic selection for enhanced fruit flavor. Proc. Natl. Acad. Sci. 119 , e2115865119 (2022).
Fritz, F., Preissner, R. & Banerjee, P. VirtualTaste: a web server for the prediction of organoleptic properties of chemical compounds. Nucleic Acids Res 49 , W679–W684 (2021).
Tuwani, R., Wadhwa, S. & Bagler, G. BitterSweet: Building machine learning models for predicting the bitter and sweet taste of small molecules. Sci. Rep. 9 , 1–13 (2019).
Dagan-Wiener, A. et al. Bitter or not? BitterPredict, a tool for predicting taste from chemical structure. Sci. Rep. 7 , 1–13 (2017).
Pallante, L. et al. Toward a general and interpretable umami taste predictor using a multi-objective machine learning approach. Sci. Rep. 12 , 1–11 (2022).
Malavolta, M. et al. A survey on computational taste predictors. Eur. Food Res. Technol. 248 , 2215–2235 (2022).
Lee, B. K. et al. A principal odor map unifies diverse tasks in olfactory perception. Science 381 , 999–1006 (2023).
Mayhew, E. J. et al. Transport features predict if a molecule is odorous. Proc. Natl. Acad. Sci. 119 , e2116576119 (2022).
Niu, Y. et al. Sensory evaluation of the synergism among ester odorants in light aroma-type liquor by odor threshold, aroma intensity and flash GC electronic nose. Food Res. Int. 113 , 102–114 (2018).
Yu, P., Low, M. Y. & Zhou, W. Design of experiments and regression modelling in food flavour and sensory analysis: A review. Trends Food Sci. Technol. 71 , 202–215 (2018).
Oladokun, O. et al. The impact of hop bitter acid and polyphenol profiles on the perceived bitterness of beer. Food Chem. 205 , 212–220 (2016).
Linforth, R., Cabannes, M., Hewson, L., Yang, N. & Taylor, A. Effect of fat content on flavor delivery during consumption: An in vivo model. J. Agric. Food Chem. 58 , 6905–6911 (2010).
Guo, S., Na Jom, K. & Ge, Y. Influence of roasting condition on flavor profile of sunflower seeds: A flavoromics approach. Sci. Rep. 9 , 11295 (2019).
Ren, Q. et al. The changes of microbial community and flavor compound in the fermentation process of Chinese rice wine using Fagopyrum tataricum grain as feedstock. Sci. Rep. 9 , 3365 (2019).
Hastie, T., Friedman, J. & Tibshirani, R. The Elements of Statistical Learning. (Springer, New York, NY). https://doi.org/10.1007/978-0-387-21606-5 (2001).
Dietz, C., Cook, D., Huismann, M., Wilson, C. & Ford, R. The multisensory perception of hop essential oil: a review. J. Inst. Brew. 126 , 320–342 (2020).
CAS Google Scholar
Roncoroni, Miguel & Verstrepen, Kevin Joan. Belgian Beer: Tested and Tasted. (Lannoo, 2018).
Meilgaard, M. Flavor chemistry of beer: Part II: Flavor and threshold of 239 aroma volatiles. in (1975).
Bokulich, N. A. & Bamforth, C. W. The microbiology of malting and brewing. Microbiol. Mol. Biol. Rev. MMBR 77 , 157–172 (2013).
Dzialo, M. C., Park, R., Steensels, J., Lievens, B. & Verstrepen, K. J. Physiology, ecology and industrial applications of aroma formation in yeast. FEMS Microbiol. Rev. 41 , S95–S128 (2017).
Article PubMed PubMed Central Google Scholar
Datta, A. et al. Computer-aided food engineering. Nat. Food 3 , 894–904 (2022).
American Society of Brewing Chemists. Beer Methods. (American Society of Brewing Chemists, St. Paul, MN, U.S.A.).
Olaniran, A. O., Hiralal, L., Mokoena, M. P. & Pillay, B. Flavour-active volatile compounds in beer: production, regulation and control. J. Inst. Brew. 123 , 13–23 (2017).
Verstrepen, K. J. et al. Flavor-active esters: Adding fruitiness to beer. J. Biosci. Bioeng. 96 , 110–118 (2003).
Meilgaard, M. C. Flavour chemistry of beer. part I: flavour interaction between principal volatiles. Master Brew. Assoc. Am. Tech. Q 12 , 107–117 (1975).
Briggs, D. E., Boulton, C. A., Brookes, P. A. & Stevens, R. Brewing 227–254. (Woodhead Publishing). https://doi.org/10.1533/9781855739062.227 (2004).
Bossaert, S., Crauwels, S., De Rouck, G. & Lievens, B. The power of sour - A review: Old traditions, new opportunities. BrewingScience 72 , 78–88 (2019).
Google Scholar
Verstrepen, K. J. et al. Flavor active esters: Adding fruitiness to beer. J. Biosci. Bioeng. 96 , 110–118 (2003).
Snauwaert, I. et al. Microbial diversity and metabolite composition of Belgian red-brown acidic ales. Int. J. Food Microbiol. 221 , 1–11 (2016).
Spitaels, F. et al. The microbial diversity of traditional spontaneously fermented lambic beer. PLoS ONE 9 , e95384 (2014).
Blanco, C. A., Andrés-Iglesias, C. & Montero, O. Low-alcohol Beers: Flavor Compounds, Defects, and Improvement Strategies. Crit. Rev. Food Sci. Nutr. 56 , 1379–1388 (2016).
Jackowski, M. & Trusek, A. Non-Alcohol. beer Prod. – Overv. 20 , 32–38 (2018).
Takoi, K. et al. The contribution of geraniol metabolism to the citrus flavour of beer: Synergy of geraniol and β-citronellol under coexistence with excess linalool. J. Inst. Brew. 116 , 251–260 (2010).
Kroeze, J. H. & Bartoshuk, L. M. Bitterness suppression as revealed by split-tongue taste stimulation in humans. Physiol. Behav. 35 , 779–783 (1985).
Mennella, J. A. et al. A spoonful of sugar helps the medicine go down”: Bitter masking bysucrose among children and adults. Chem. Senses 40 , 17–25 (2015).
Wietstock, P., Kunz, T., Perreira, F. & Methner, F.-J. Metal chelation behavior of hop acids in buffered model systems. BrewingScience 69 , 56–63 (2016).
Sancho, D., Blanco, C. A., Caballero, I. & Pascual, A. Free iron in pale, dark and alcohol-free commercial lager beers. J. Sci. Food Agric. 91 , 1142–1147 (2011).
Rodrigues, H. & Parr, W. V. Contribution of cross-cultural studies to understanding wine appreciation: A review. Food Res. Int. 115 , 251–258 (2019).
Korneva, E. & Blockeel, H. Towards better evaluation of multi-target regression models. in ECML PKDD 2020 Workshops (eds. Koprinska, I. et al.) 353–362 (Springer International Publishing, Cham, 2020). https://doi.org/10.1007/978-3-030-65965-3_23 .
Gastón Ares. Mathematical and Statistical Methods in Food Science and Technology. (Wiley, 2013).
Grinsztajn, L., Oyallon, E. & Varoquaux, G. Why do tree-based models still outperform deep learning on tabular data? Preprint at http://arxiv.org/abs/2207.08815 (2022).
Gries, S. T. Statistics for Linguistics with R: A Practical Introduction. in Statistics for Linguistics with R (De Gruyter Mouton, 2021). https://doi.org/10.1515/9783110718256 .
Lundberg, S. M. et al. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2 , 56–67 (2020).
Ickes, C. M. & Cadwallader, K. R. Effects of ethanol on flavor perception in alcoholic beverages. Chemosens. Percept. 10 , 119–134 (2017).
Kato, M. et al. Influence of high molecular weight polypeptides on the mouthfeel of commercial beer. J. Inst. Brew. 127 , 27–40 (2021).
Wauters, R. et al. Novel Saccharomyces cerevisiae variants slow down the accumulation of staling aldehydes and improve beer shelf-life. Food Chem. 398 , 1–11 (2023).
Li, H., Jia, S. & Zhang, W. Rapid determination of low-level sulfur compounds in beer by headspace gas chromatography with a pulsed flame photometric detector. J. Am. Soc. Brew. Chem. 66 , 188–191 (2008).
Dercksen, A., Laurens, J., Torline, P., Axcell, B. C. & Rohwer, E. Quantitative analysis of volatile sulfur compounds in beer using a membrane extraction interface. J. Am. Soc. Brew. Chem. 54 , 228–233 (1996).
Molnar, C. Interpretable Machine Learning: A Guide for Making Black-Box Models Interpretable. (2020).
Zhao, Q. & Hastie, T. Causal interpretations of black-box models. J. Bus. Econ. Stat. Publ. Am. Stat. Assoc. 39 , 272–281 (2019).
Article MathSciNet Google Scholar
Hastie, T., Tibshirani, R. & Friedman, J. The Elements of Statistical Learning. (Springer, 2019).
Labrado, D. et al. Identification by NMR of key compounds present in beer distillates and residual phases after dealcoholization by vacuum distillation. J. Sci. Food Agric. 100 , 3971–3978 (2020).
Lusk, L. T., Kay, S. B., Porubcan, A. & Ryder, D. S. Key olfactory cues for beer oxidation. J. Am. Soc. Brew. Chem. 70 , 257–261 (2012).
Gonzalez Viejo, C., Torrico, D. D., Dunshea, F. R. & Fuentes, S. Development of artificial neural network models to assess beer acceptability based on sensory properties using a robotic pourer: A comparative model approach to achieve an artificial intelligence system. Beverages 5 , 33 (2019).
Gonzalez Viejo, C., Fuentes, S., Torrico, D. D., Godbole, A. & Dunshea, F. R. Chemical characterization of aromas in beer and their effect on consumers liking. Food Chem. 293 , 479–485 (2019).
Gilbert, J. L. et al. Identifying breeding priorities for blueberry flavor using biochemical, sensory, and genotype by environment analyses. PLOS ONE 10 , 1–21 (2015).
Goulet, C. et al. Role of an esterase in flavor volatile variation within the tomato clade. Proc. Natl. Acad. Sci. 109 , 19009–19014 (2012).
Article ADS CAS PubMed PubMed Central Google Scholar
Borisov, V. et al. Deep Neural Networks and Tabular Data: A Survey. IEEE Trans. Neural Netw. Learn. Syst. 1–21 https://doi.org/10.1109/TNNLS.2022.3229161 (2022).
Statista. Statista Consumer Market Outlook: Beer - Worldwide.
Seitz, H. K. & Stickel, F. Molecular mechanisms of alcoholmediated carcinogenesis. Nat. Rev. Cancer 7 , 599–612 (2007).
Voordeckers, K. et al. Ethanol exposure increases mutation rate through error-prone polymerases. Nat. Commun. 11 , 3664 (2020).
Goelen, T. et al. Bacterial phylogeny predicts volatile organic compound composition and olfactory response of an aphid parasitoid. Oikos 129 , 1415–1428 (2020).
Article ADS Google Scholar
Reher, T. et al. Evaluation of hop (Humulus lupulus) as a repellent for the management of Drosophila suzukii. Crop Prot. 124 , 104839 (2019).
Stein, S. E. An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectrom. 10 , 770–781 (1999).
American Society of Brewing Chemists. Sensory Analysis Methods. (American Society of Brewing Chemists, St. Paul, MN, U.S.A., 1992).
McAuley, J., Leskovec, J. & Jurafsky, D. Learning Attitudes and Attributes from Multi-Aspect Reviews. Preprint at https://doi.org/10.48550/arXiv.1210.3926 (2012).
Meilgaard, M. C., Carr, B. T. & Carr, B. T. Sensory Evaluation Techniques. (CRC Press, Boca Raton). https://doi.org/10.1201/b16452 (2014).
Schreurs, M. et al. Data from: Predicting and improving complex beer flavor through machine learning. Zenodo https://doi.org/10.5281/zenodo.10653704 (2024).
Download references
Acknowledgements
We thank all lab members for their discussions and thank all tasting panel members for their contributions. Special thanks go out to Dr. Karin Voordeckers for her tremendous help in proofreading and improving the manuscript. M.S. was supported by a Baillet-Latour fellowship, L.C. acknowledges financial support from KU Leuven (C16/17/006), F.A.T. was supported by a PhD fellowship from FWO (1S08821N). Research in the lab of K.J.V. is supported by KU Leuven, FWO, VIB, VLAIO and the Brewing Science Serves Health Fund. Research in the lab of T.W. is supported by FWO (G.0A51.15) and KU Leuven (C16/17/006).
Author information
These authors contributed equally: Michiel Schreurs, Supinya Piampongsant, Miguel Roncoroni.
Authors and Affiliations
VIB—KU Leuven Center for Microbiology, Gaston Geenslaan 1, B-3001, Leuven, Belgium
Michiel Schreurs, Supinya Piampongsant, Miguel Roncoroni, Lloyd Cool, Beatriz Herrera-Malaver, Florian A. Theßeling & Kevin J. Verstrepen
CMPG Laboratory of Genetics and Genomics, KU Leuven, Gaston Geenslaan 1, B-3001, Leuven, Belgium
Leuven Institute for Beer Research (LIBR), Gaston Geenslaan 1, B-3001, Leuven, Belgium
Laboratory of Socioecology and Social Evolution, KU Leuven, Naamsestraat 59, B-3000, Leuven, Belgium
Lloyd Cool, Christophe Vanderaa & Tom Wenseleers
VIB Bioinformatics Core, VIB, Rijvisschestraat 120, B-9052, Ghent, Belgium
Łukasz Kreft & Alexander Botzki
AB InBev SA/NV, Brouwerijplein 1, B-3000, Leuven, Belgium
Philippe Malcorps & Luk Daenen
You can also search for this author in PubMed Google Scholar
Contributions
S.P., M.S. and K.J.V. conceived the experiments. S.P., M.S. and K.J.V. designed the experiments. S.P., M.S., M.R., B.H. and F.A.T. performed the experiments. S.P., M.S., L.C., C.V., L.K., A.B., P.M., L.D., T.W. and K.J.V. contributed analysis ideas. S.P., M.S., L.C., C.V., T.W. and K.J.V. analyzed the data. All authors contributed to writing the manuscript.
Corresponding author
Correspondence to Kevin J. Verstrepen .
Ethics declarations
Competing interests.
K.J.V. is affiliated with bar.on. The other authors declare no competing interests.
Peer review
Peer review information.
Nature Communications thanks Florian Bauer, Andrew John Macintosh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information, peer review file, description of additional supplementary files, supplementary data 1, supplementary data 2, supplementary data 3, supplementary data 4, supplementary data 5, supplementary data 6, supplementary data 7, reporting summary, source data, source data, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Schreurs, M., Piampongsant, S., Roncoroni, M. et al. Predicting and improving complex beer flavor through machine learning. Nat Commun 15 , 2368 (2024). https://doi.org/10.1038/s41467-024-46346-0
Download citation
Received : 30 October 2023
Accepted : 21 February 2024
Published : 26 March 2024
DOI : https://doi.org/10.1038/s41467-024-46346-0
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.

IMAGES
VIDEO
COMMENTS
After you get enough feedback and decide on the journal you will submit to, the process of real writing begins. Copy your outline into a separate file and expand on each of the points, adding data and elaborating on the details. When you create the first draft, do not succumb to the temptation of editing.
Structure for writing a scientific research proposal in biotechnology. The aim or goal and objective of the biotechnology research proposal should give a broad indication of the expected research outcome and the hypothesis to be tested can also be the aim of your study. The objective can be categorized as primary and secondary according to the ...
Tell a story . Condense large amounts of information and Simplify complex findings. Convince readers of your findings (by showing data quality or experimental design, apparatus) Focus attention (e.g., relationship between values) Promote thinking and discussion .
Language and manuscript preparation services. Let one of our experts assist you with getting your manuscript and language into shape - our services cover: English language improvement. scientific in-depth editing and strategic advice. figure and tables formatting. manuscript formatting to match your target journal.
The Introduction and Discussion are like bookends around your Methods and Results. They integrate your original findings with the literature and current knowledge. Context, Justification, and Focus. addressed in the Introduction are echoed in the Discussion section: Focus = Summary of findings.
A clear format will ensure that your research paper is understood by your readers. Follow: 1. Context — your introduction. 2. Content — your results. 3. Conclusion — your discussion. Plan ...
Step 3: Collect enough data. Collect or continue to collect data for your study until the amount of data required for your target journal is met. Drafting a list of the tables and figures - and even the figure legends - before you even start your experiment can help streamline your data collection process.
Functions of the Title. The title of a biomedical scientific paper has two main functions ( 18, 19 ): (1) to present the main topic or the message of the paper (the answer to the question) and (2) to attract potential readers and evoke their interest to read the paper. In fact, the title tells the readers what the paper is all about ( 6, 19 ).
Seven tips on crafting papers. (1) The art of positive visualization: start with the figure set. I learned early in my career from eminent colleagues that one good strategy can be to start with the figure set… even before we have the full and final set of data for the paper! It's a great aid for crafting the logic and flow of the paper.
In the following part of this paper the different sections of a manuscript will be dealt with in the order they are presented in the final document. Go to: Title, keywords and abstract. The title is determinant for the indexing process of the article and greatly contributes to the visibility of the paper.
advice on writing about their work and getting published. ... submitting, and publishing scientific papers is now largely elec-tronic, and the book has been revised to reflect this. New material features more information on including supplementary material online, using ref- ... and Biotechnology at Texas A&M University.
Lectures on Scientific Communications. 1: Basic Scientific Communication ( PDF) 2: How to Review the Literature ( PDF) 3: How To Write a Research Proposal ( PDF) 4: Preparing Effective Oral Presentations ( PDF) 5: How to Write a Mini Literature Review ( PDF) 6: How to Write a Research Paper I: Illustrations ( PDF - 1.2 MB)
In Biotechnology, students are trained in a broad range of basic sciences as a foundation for many important real-world applications. A major part of this training is participation in research. All Biotechnology majors participate in at least 3 credit hours of research. Students have a large number of research laboratories across all the ...
In this latest addition of biotechnology literature analysis, we aimed to unveil the latest trends (since 2017) in biotechnology research. By analyzing the research literature, we identified the latest popular research themes, major contributors in terms of institutions, countries/regions, and journals. 2. Materials and methods.
Describe the specific research methods you plan to use. Common methods in biotechnology include: Experiments (e.g., cell culture, protein purification) Computational modeling. Surveys of relevant specialists. Literature reviews and meta-analyses of existing data. Justify why your chosen methods are the most appropriate for answering your ...
Biotechnology Research Paper Topics. This collection of biotechnology research paper topics provides the list of 10 potential topics for research papers and overviews the history of biotechnology. The term biotechnology came into popular use around 1980 and was understood to mean the industrial use of microorganisms to make goods and services ...
To submit your manuscript, register and log in to the submission website. Once you have registered, click here to go to the submission form for BioTech. All co-authors can see the manuscript details in the submission system, if they register and log in using the e-mail address provided during manuscript submission.
How to write and give a talk or present a paper to any professional group. • How to write and give a talk or present a paper in English to an ESPEN audience. • How to match the style of the talk to the level of the audience. • How to improve in two areas - from poor speaker with poor slides to good speaker with good slides.
Citation is the process of citing the original research paper in which the idea or methods used for composing a research proposal. And it is mandatory for any scientific research articles. In summary, a research proposal in biotechnology should communicate the researcher's knowledge on the project, methods and explain the need for the study ...
A Help with english homework shares 15 trending biotechnology research topics. The interrelation between the consumption of iron-folic acid during pregnancy and its effect on the complete health ...
Research Paper 01 Oct 1992. ... Nature Biotechnology (Nat Biotechnol) ISSN 1546-1696 (online) ISSN 1087-0156 (print) nature.com sitemap. About Nature Portfolio. About us ...
Biotechnology is a broad discipline in which biological processes, organisms, cells or cellular components are exploited to develop new technologies. New tools and products developed by ...
Rule 2: Write the introduction: describe the purpose (the why) An essential aspect of any SOP is introduction and purpose forming. The introduction section specifies the need and capability of the procedures for the research environment in which the process is being established. Identify the specific reason you decided to write your SOP.
🔥Join me for my Certification Course on 'A-Z of Research Writing & Presentation' 😃: https://wiseupcommunications.com/course/research-writing/In this video ...
Follow these tips to produce stronger writing that stands out on the web even in the age of AI and ChatGPT. Whether you're a student, a journalist, or a business professional, knowing how to do ...
Abstract. The perception and appreciation of food flavor depends on many interacting chemical compounds and external factors, and therefore proves challenging to understand and predict. Here, we ...