- Research article
- Open access
- Published: 21 December 2019

Comparing different supervised machine learning algorithms for disease prediction
- Shahadat Uddin ORCID: orcid.org/0000-0003-0091-6919 1 ,
- Arif Khan 1 , 2 ,
- Md Ekramul Hossain 1 &
- Mohammad Ali Moni 3
BMC Medical Informatics and Decision Making volume 19 , Article number: 281 ( 2019 ) Cite this article
119k Accesses
666 Citations
13 Altmetric
Metrics details
Supervised machine learning algorithms have been a dominant method in the data mining field. Disease prediction using health data has recently shown a potential application area for these methods. This study aims to identify the key trends among different types of supervised machine learning algorithms, and their performance and usage for disease risk prediction.
In this study, extensive research efforts were made to identify those studies that applied more than one supervised machine learning algorithm on single disease prediction. Two databases (i.e., Scopus and PubMed) were searched for different types of search items. Thus, we selected 48 articles in total for the comparison among variants supervised machine learning algorithms for disease prediction.
We found that the Support Vector Machine (SVM) algorithm is applied most frequently (in 29 studies) followed by the Naïve Bayes algorithm (in 23 studies). However, the Random Forest (RF) algorithm showed superior accuracy comparatively. Of the 17 studies where it was applied, RF showed the highest accuracy in 9 of them, i.e., 53%. This was followed by SVM which topped in 41% of the studies it was considered.
This study provides a wide overview of the relative performance of different variants of supervised machine learning algorithms for disease prediction. This important information of relative performance can be used to aid researchers in the selection of an appropriate supervised machine learning algorithm for their studies.
Peer Review reports
Machine learning algorithms employ a variety of statistical, probabilistic and optimisation methods to learn from past experience and detect useful patterns from large, unstructured and complex datasets [ 1 ]. These algorithms have a wide range of applications, including automated text categorisation [ 2 ], network intrusion detection [ 3 ], junk e-mail filtering [ 4 ], detection of credit card fraud [ 5 ], customer purchase behaviour detection [ 6 ], optimising manufacturing process [ 7 ] and disease modelling [ 8 ]. Most of these applications have been implemented using supervised variants [ 4 , 5 , 8 ] of the machine learning algorithms rather than unsupervised ones. In the supervised variant, a prediction model is developed by learning a dataset where the label is known and accordingly the outcome of unlabelled examples can be predicted [ 9 ].
The scope of this research is primarily on the performance analysis of disease prediction approaches using different variants of supervised machine learning algorithms. Disease prediction and in a broader context, medical informatics, have recently gained significant attention from the data science research community in recent years. This is primarily due to the wide adaptation of computer-based technology into the health sector in different forms (e.g., electronic health records and administrative data) and subsequent availability of large health databases for researchers. These electronic data are being utilised in a wide range of healthcare research areas such as the analysis of healthcare utilisation [ 10 ], measuring performance of a hospital care network [ 11 ], exploring patterns and cost of care [ 12 ], developing disease risk prediction model [ 13 , 14 ], chronic disease surveillance [ 15 ], and comparing disease prevalence and drug outcomes [ 16 ]. Our research focuses on the disease risk prediction models involving machine learning algorithms (e.g., support vector machine, logistic regression and artificial neural network), specifically - supervised learning algorithms. Models based on these algorithms use labelled training data of patients for training [ 8 , 17 , 18 ]. For the test set, patients are classified into several groups such as low risk and high risk.
Given the growing applicability and effectiveness of supervised machine learning algorithms on predictive disease modelling, the breadth of research still seems progressing. Specifically, we found little research that makes a comprehensive review of published articles employing different supervised learning algorithms for disease prediction. Therefore, this research aims to identify key trends among different types of supervised machine learning algorithms, their performance accuracies and the types of diseases being studied. In addition, the advantages and limitations of different supervised machine learning algorithms are summarised. The results of this study will help the scholars to better understand current trends and hotspots of disease prediction models using supervised machine learning algorithms and formulate their research goals accordingly.
In making comparisons among different supervised machine learning algorithms, this study reviewed, by following the PRISMA guidelines [ 19 ], existing studies from the literature that used such algorithms for disease prediction. More specifically, this article considered only those studies that used more than one supervised machine learning algorithm for a single disease prediction in the same research setting. This made the principal contribution of this study (i.e., comparison among different supervised machine learning algorithms) more accurate and comprehensive since the comparison of the performance of a single algorithm across different study settings can be biased and generate erroneous results [ 20 ].
Traditionally, standard statistical methods and doctor’s intuition, knowledge and experience had been used for prognosis and disease risk prediction. This practice often leads to unwanted biases, errors and high expenses, and negatively affects the quality of service provided to patients [ 21 ]. With the increasing availability of electronic health data, more robust and advanced computational approaches such as machine learning have become more practical to apply and explore in disease prediction area. In the literature, most of the related studies utilised one or more machine learning algorithms for a particular disease prediction. For this reason, the performance comparison of different supervised machine learning algorithms for disease prediction is the primary focus of this study.
In the following sections, we discuss different variants of supervised machine learning algorithm, followed by presenting the methods of this study. In the subsequent sections, we present the results and discussion of the study.
- Supervised machine learning algorithm
At its most basic sense, machine learning uses programmed algorithms that learn and optimise their operations by analysing input data to make predictions within an acceptable range. With the feeding of new data, these algorithms tend to make more accurate predictions. Although there are some variations of how to group machine learning algorithms they can be divided into three broad categories according to their purposes and the way the underlying machine is being taught. These three categories are: supervised, unsupervised and semi-supervised.
In supervised machine learning algorithms, a labelled training dataset is used first to train the underlying algorithm. This trained algorithm is then fed on the unlabelled test dataset to categorise them into similar groups. Using an abstract dataset for three diabetic patients, Fig. 1 shows an illustration about how supervised machine learning algorithms work to categorise diabetic and non-diabetic patients. Supervised learning algorithms suit well with two types of problems: classification problems; and regression problems. In classification problems, the underlying output variable is discrete. This variable is categorised into different groups or categories, such as ‘red’ or ‘black’, or it could be ‘diabetic’ and ‘non-diabetic’. The corresponding output variable is a real value in regression problems, such as the risk of developing cardiovascular disease for an individual. In the following subsections, we briefly describe the commonly used supervised machine learning algorithms for disease prediction.
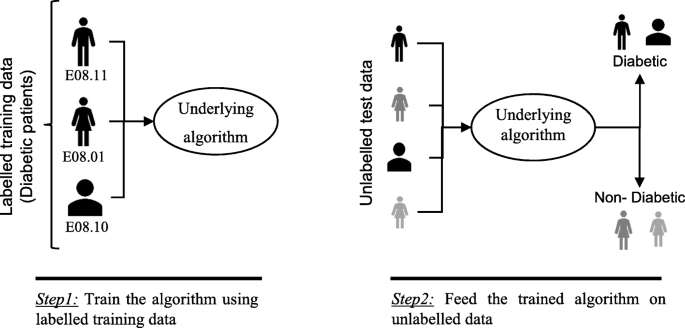
An illustration of how supervised machine learning algorithms work to categorise diabetic and non-diabetic patients based on abstract data
Logistic regression
Logistic regression (LR) is a powerful and well-established method for supervised classification [ 22 ]. It can be considered as an extension of ordinary regression and can model only a dichotomous variable which usually represents the occurrence or non-occurrence of an event. LR helps in finding the probability that a new instance belongs to a certain class. Since it is a probability, the outcome lies between 0 and 1. Therefore, to use the LR as a binary classifier, a threshold needs to be assigned to differentiate two classes. For example, a probability value higher than 0.50 for an input instance will classify it as ‘class A’; otherwise, ‘class B’. The LR model can be generalised to model a categorical variable with more than two values. This generalised version of LR is known as the multinomial logistic regression.
Support vector machine
Support vector machine (SVM) algorithm can classify both linear and non-linear data. It first maps each data item into an n-dimensional feature space where n is the number of features. It then identifies the hyperplane that separates the data items into two classes while maximising the marginal distance for both classes and minimising the classification errors [ 23 ]. The marginal distance for a class is the distance between the decision hyperplane and its nearest instance which is a member of that class. More formally, each data point is plotted first as a point in an n-dimension space (where n is the number of features) with the value of each feature being the value of a specific coordinate. To perform the classification, we then need to find the hyperplane that differentiates the two classes by the maximum margin. Figure 2 provides a simplified illustration of an SVM classifier.
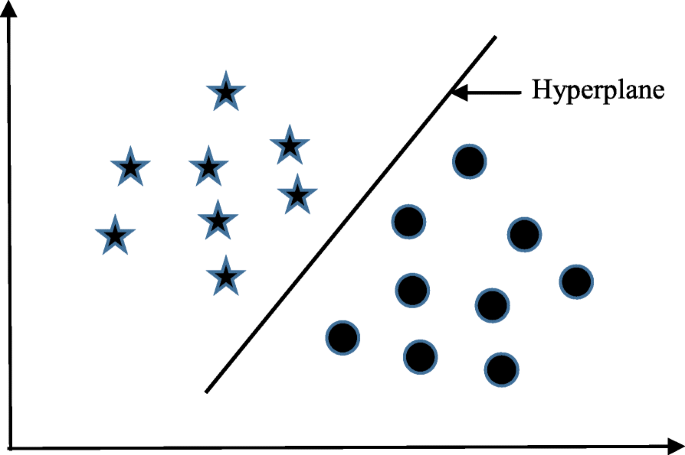
A simplified illustration of how the support vector machine works. The SVM has identified a hyperplane (actually a line) which maximises the separation between the ‘star’ and ‘circle’ classes
Decision tree
Decision tree (DT) is one of the earliest and prominent machine learning algorithms. A decision tree models the decision logics i.e., tests and corresponds outcomes for classifying data items into a tree-like structure. The nodes of a DT tree normally have multiple levels where the first or top-most node is called the root node. All internal nodes (i.e., nodes having at least one child) represent tests on input variables or attributes. Depending on the test outcome, the classification algorithm branches towards the appropriate child node where the process of test and branching repeats until it reaches the leaf node [ 24 ]. The leaf or terminal nodes correspond to the decision outcomes. DTs have been found easy to interpret and quick to learn, and are a common component to many medical diagnostic protocols [ 25 ]. When traversing the tree for the classification of a sample, the outcomes of all tests at each node along the path will provide sufficient information to conjecture about its class. An illustration of an DT with its elements and rules is depicted in Fig. 3 .
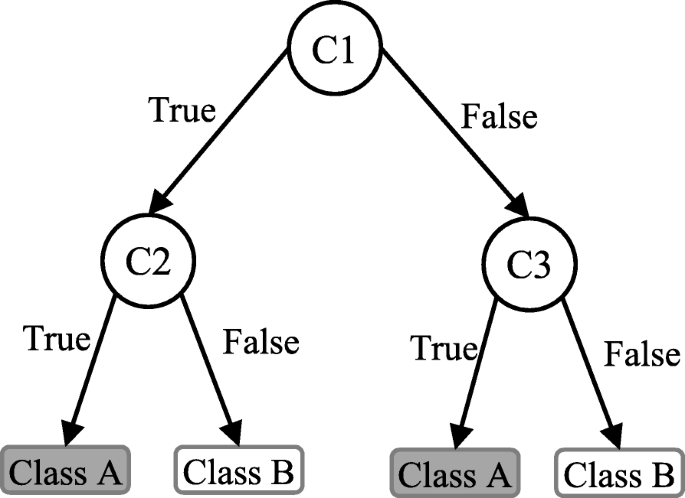
An illustration of a Decision tree. Each variable (C1, C2, and C3) is represented by a circle and the decision outcomes (Class A and Class B) are shown by rectangles. In order to successfully classify a sample to a class, each branch is labelled with either ‘True’ or ‘False’ based on the outcome value from the test of its ancestor node
Random forest
A random forest (RF) is an ensemble classifier and consisting of many DTs similar to the way a forest is a collection of many trees [ 26 ]. DTs that are grown very deep often cause overfitting of the training data, resulting a high variation in classification outcome for a small change in the input data. They are very sensitive to their training data, which makes them error-prone to the test dataset. The different DTs of an RF are trained using the different parts of the training dataset. To classify a new sample, the input vector of that sample is required to pass down with each DT of the forest. Each DT then considers a different part of that input vector and gives a classification outcome. The forest then chooses the classification of having the most ‘votes’ (for discrete classification outcome) or the average of all trees in the forest (for numeric classification outcome). Since the RF algorithm considers the outcomes from many different DTs, it can reduce the variance resulted from the consideration of a single DT for the same dataset. Figure 4 shows an illustration of the RF algorithm.
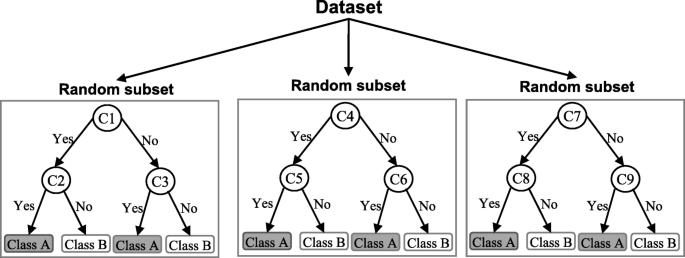
An illustration of a Random forest which consists of three different decision trees. Each of those three decision trees was trained using a random subset of the training data
Naïve Bayes
Naïve Bayes (NB) is a classification technique based on the Bayes’ theorem [ 27 ]. This theorem can describe the probability of an event based on the prior knowledge of conditions related to that event. This classifier assumes that a particular feature in a class is not directly related to any other feature although features for that class could have interdependence among themselves [ 28 ]. By considering the task of classifying a new object (white circle) to either ‘green’ class or ‘red’ class, Fig. 5 provides an illustration about how the NB technique works. According to this figure, it is reasonable to believe that any new object is twice as likely to have ‘green’ membership rather than ‘red’ since there are twice as many ‘green’ objects (40) as ‘red’. In the Bayesian analysis, this belief is known as the prior probability. Therefore, the prior probabilities of ‘green’ and ‘red’ are 0.67 (40 ÷ 60) and 0.33 (20 ÷ 60), respectively. Now to classify the ‘white’ object, we need to draw a circle around this object which encompasses several points (to be chosen prior) irrespective of their class labels. Four points (three ‘red’ and one ‘green) were considered in this figure. Thus, the likelihood of ‘white’ given ‘green’ is 0.025 (1 ÷ 40) and the likelihood of ‘white’ given ‘red’ is 0.15 (3 ÷ 20). Although the prior probability indicates that the new ‘white’ object is more likely to have ‘green’ membership, the likelihood shows that it is more likely to be in the ‘red’ class. In the Bayesian analysis, the final classifier is produced by combining both sources of information (i.e., prior probability and likelihood value). The ‘multiplication’ function is used to combine these two types of information and the product is called the ‘posterior’ probability. Finally, the posterior probability of ‘white’ being ‘green’ is 0.017 (0.67 × 0.025) and the posterior probability of ‘white’ being ‘red’ is 0.049 (0.33 × 0.15). Thus, the new ‘white’ object should be classified as a member of the ‘red’ class according to the NB technique.
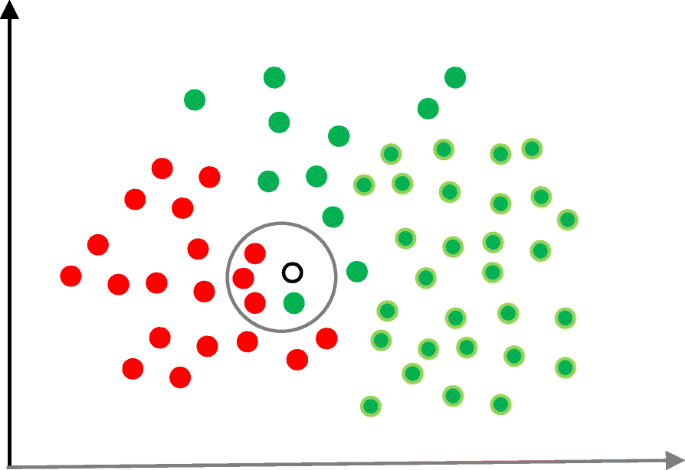
An illustration of the Naïve Bayes algorithm. The ‘white’ circle is the new sample instance which needs to be classified either to ‘red’ class or ‘green’ class
K-nearest neighbour
The K-nearest neighbour (KNN) algorithm is one of the simplest and earliest classification algorithms [ 29 ]. It can be thought a simpler version of an NB classifier. Unlike the NB technique, the KNN algorithm does not require to consider probability values. The ‘ K ’ is the KNN algorithm is the number of nearest neighbours considered to take ‘vote’ from. The selection of different values for ‘ K ’ can generate different classification results for the same sample object. Figure 6 shows an illustration of how the KNN works to classify a new object. For K = 3 , the new object (star) is classified as ‘black’; however, it has been classified as ‘red’ when K = 5 .
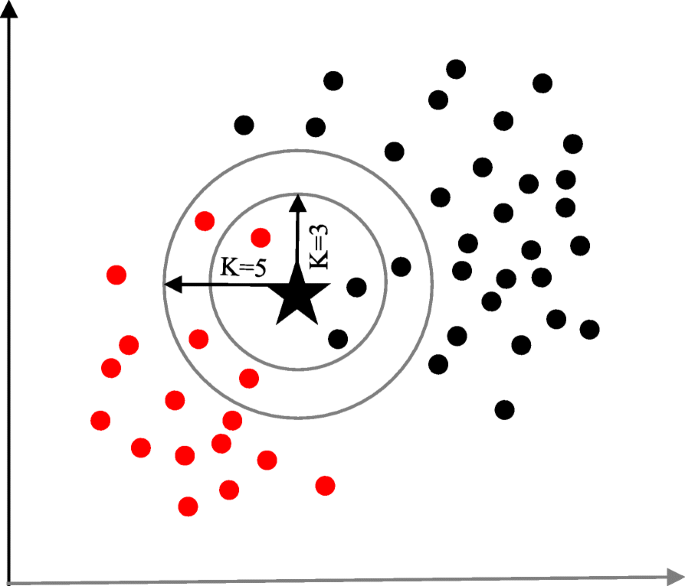
A simplified illustration of the K-nearest neighbour algorithm. When K = 3, the sample object (‘star’) is classified as ‘black’ since it gets more ‘vote’ from the ‘black’ class. However, for K = 5 the same sample object is classified as ‘red’ since it now gets more ‘vote’ from the ‘red’ class
Artificial neural network
Artificial neural networks (ANNs) are a set of machine learning algorithms which are inspired by the functioning of the neural networks of human brain. They were first proposed by McCulloch and Pitts [ 30 ] and later popularised by the works of Rumelhart et al. in the 1980s [ 31 ].. In the biological brain, neurons are connected to each other through multiple axon junctions forming a graph like architecture. These interconnections can be rewired (e.g., through neuroplasticity) that helps to adapt, process and store information. Likewise, ANN algorithms can be represented as an interconnected group of nodes. The output of one node goes as input to another node for subsequent processing according to the interconnection. Nodes are normally grouped into a matrix called layer depending on the transformation they perform. Apart from the input and output layer, there can be one or more hidden layers in an ANN framework. Nodes and edges have weights that enable to adjust signal strengths of communication which can be amplified or weakened through repeated training. Based on the training and subsequent adaption of the matrices, node and edge weights, ANNs can make a prediction for the test data. Figure 7 shows an illustration of an ANN (with two hidden layers) with its interconnected group of nodes.
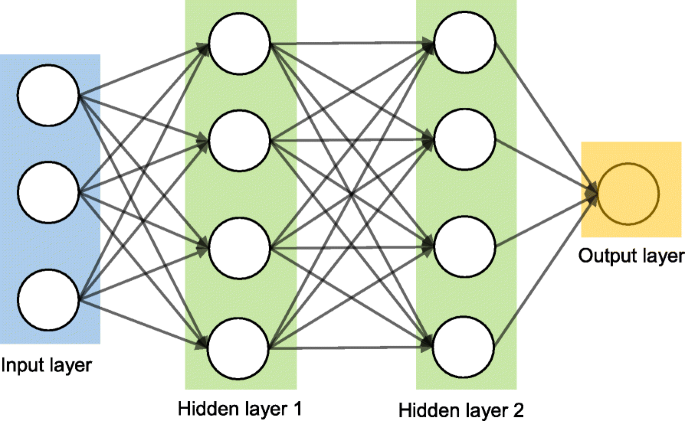
An illustration of the artificial neural network structure with two hidden layers. The arrows connect the output of nodes from one layer to the input of nodes of another layer
Data source and data extraction
Extensive research efforts were made to identify articles employing more than one supervised machine learning algorithm for disease prediction. Two databases were searched (October 2018): Scopus and PubMed. Scopus is an online bibliometric database developed by Elsevier. It has been chosen because of its high level of accuracy and consistency [ 32 ]. PubMed is a free publication search engine and incorporates citation information mostly for biomedical and life science literature. It comprises more than 28 million citations from MEDLINE, life science journals and online books [ 33 ]. MEDLINE is a bibliographic database that includes bibliographic information for articles from academic journals covering medicine, nursing, pharmacy, dentistry, veterinary medicine, and health care [ 33 ].
A comprehensive search strategy was followed to find out all related articles. The search terms that were used in this search strategy were:
“disease prediction” AND “machine learning”;
“disease prediction” AND “data mining”;
“disease risk prediction” AND “machine learning”; and
“disease risk prediction” AND “data mining”.
In scientific literature, the generic name of “machine learning” is often used for both “supervised” and “unsupervised” machine learning algorithms. On the other side, there is a close relationship between the terms “machine learning” and “data mining”, with the latter is commonly used for the former one [ 34 ]. For these reasons, we used both “machine learning” and “data mining” in the search terms although the focus of this study is on the supervised machine learning algorithm. The four search items were then considered to launch searches on the titles, abstracts and keywords of an article for both Scopus and PubMed. This resulted in 305 and 83 articles from Scopus and PubMed, respectively. After combining these two lists of articles and removing the articles written in languages other than English, we found 336 unique articles.
Since the aim of this study was to compare the performance of different supervised machine learning algorithms, the next step was to select the articles from these 336 which used more than one supervised machine learning algorithm for disease prediction. For this reason, we wrote a computer program using Python programming language [ 35 ] which checked the presence of the name of more than one supervised machine learning algorithm in the title, abstract and keyword list of each of 336 articles. It found 55 articles that used more than one supervised machine learning algorithm for the prediction of different diseases. Out of the remaining 281 articles, only 155 used one of the seven supervised machine learning algorithms considered in this study. The rest 126 used either other machine learning algorithms (e.g., unsupervised or semi-supervised) or data mining methods other than machine learning ones. ANN was found most frequently (30.32%) in the 155 articles, followed by the Naïve Bayes (19.35%).
The next step is the manual inspection of all recovered articles. We noticed that four groups of authors reported their study results in two publication outlets (i.e., book chapter, conference and journal) using the same or different titles. For these four publications, we considered the most recent one. We further excluded another three articles since the reported prediction accuracies for all supervised machine learning algorithms used in those articles are the same. For each of the remaining 48 articles, the performance outcomes of the supervised machine learning algorithms that were used for disease prediction were gathered. Two diseases were predicted in one article [ 17 ] and two algorithms were found showing the best accuracy outcomes for a disease in one article [ 36 ]. In that article, five different algorithms were used for prediction analysis. The number of publications per year has been depicted in Fig. 8 . The overall data collection procedure along with the number of articles selected for different diseases has been shown in Fig. 9 .
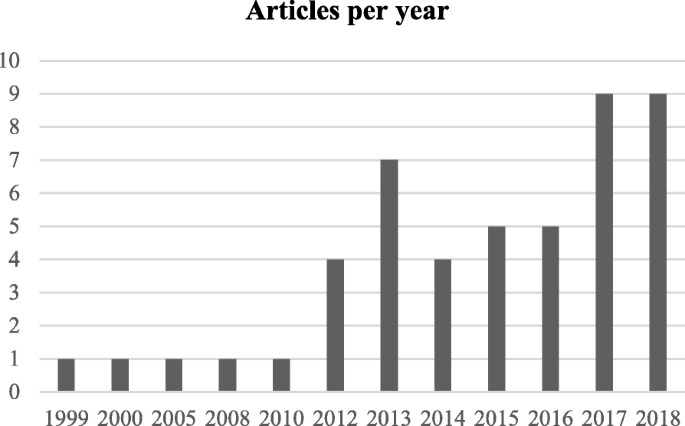
Number of articles published in different years
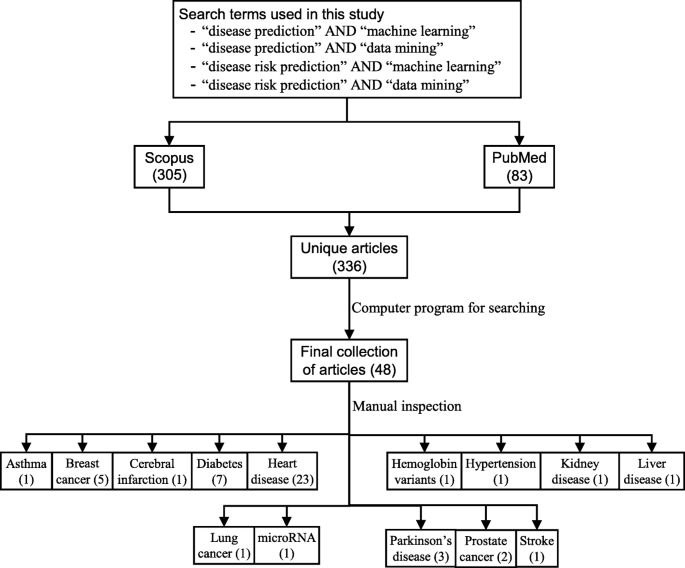
The overall data collection procedure. It also shows the number of articles considered for each disease
Figure 10 shows a comparison of the composition of initially selected 329 articles regarding the seven supervised machine learning algorithms considered in this study. ANN shows the highest percentage difference (i.e., 16%) between the 48 selected articles of this study and initially selected 155 articles that used only one supervised machine learning algorithm for disease prediction, which is followed by LR. The remaining five supervised machine learning algorithms show a percentage difference between 1 and 5.
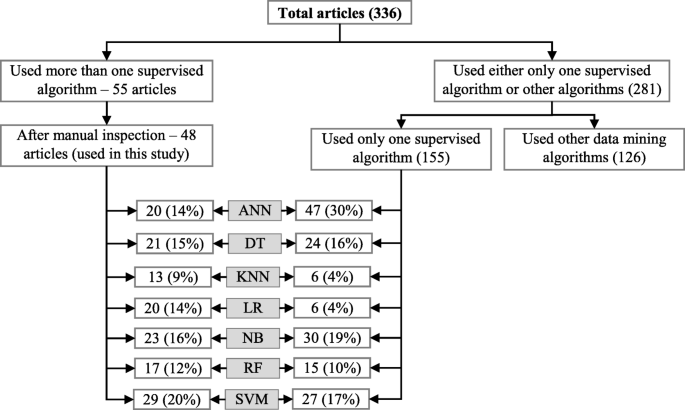
Composition of initially selected 329 articles with respect to the seven supervised learning algorithms
Classifier performance index
The diagnostic ability of classifiers has usually been determined by the confusion matrix and the receiver operating characteristic (ROC) curve [ 37 ]. In the machine learning research domain, the confusion matrix is also known as error or contingency matrix. The basic framework of the confusion matrix has been provided in Fig. 11 a. In this framework, true positives (TP) are the positive cases where the classifier correctly identified them. Similarly, true negatives (TN) are the negative cases where the classifier correctly identified them. False positives (FP) are the negative cases where the classifier incorrectly identified them as positive and the false negatives (FN) are the positive cases where the classifier incorrectly identified them as negative. The following measures, which are based on the confusion matrix, are commonly used to analyse the performance of classifiers, including those that are based on supervised machine learning algorithms.
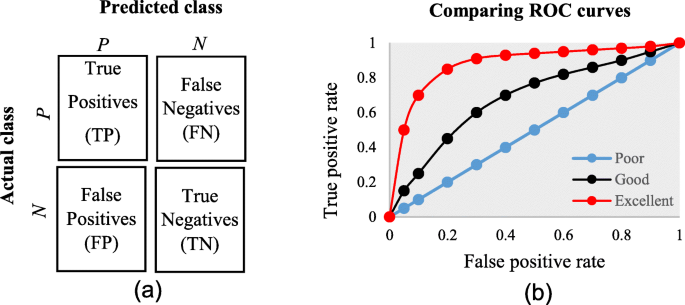
a The basic framework of the confusion matrix; and ( b ) A presentation of the ROC curve
An ROC is one of the fundamental tools for diagnostic test evaluation and is created by plotting the true positive rate against the false positive rate at various threshold settings [ 37 ]. The area under the ROC curve (AUC) is also commonly used to determine the predictability of a classifier. A higher AUC value represents the superiority of a classifier and vice versa. Figure 11 b illustrates a presentation of three ROC curves based on an abstract dataset. The area under the blue ROC curve is half of the shaded rectangle. Thus, the AUC value for this blue ROC curve is 0.5. Due to the coverage of a larger area, the AUC value for the red ROC curve is higher than that of the black ROC curve. Hence, the classifier that produced the red ROC curve shows higher predictive accuracy compared with the other two classifiers that generated the blue and red ROC curves.
There are few other measures that are also used to assess the performance of different classifiers. One such measure is the running mean square error (RMSE). For different pairs of actual and predicted values, RMSE represents the mean value of all square errors. An error is the difference between an actual and its corresponding predicted value. Another such measure is the mean absolute error (MAE). For an actual and its predicted value, MAE indicates the absolute value of their difference.
The final dataset contained 48 articles, each of which implemented more than one variant of supervised machine learning algorithms for a single disease prediction. All implemented variants were already discussed in the methods section as well as the more frequently used performance measures. Based on these, we reviewed the finally selected 48 articles in terms of the methods used, performance measures as well as the disease they targeted.
In Table 1 , names and references of the diseases and the corresponding supervised machine learning algorithms used to predict them are discussed. For each of the disease models, the better performing algorithm is also described in this table. This study considered 48 articles, which in total made the prediction for 49 diseases or conditions (one article predicted two diseases [ 17 ]). For these 49 diseases, 50 algorithms were found to show the superior accuracy. One disease has two algorithms (out of 5) that showed the same higher-level accuracies [ 36 ]. To sum up, 49 diseases were predicted in 48 articles considered in this study and 50 supervised machine learning algorithms were found to show the superior accuracy. The advantages and limitations of different supervised machine learning algorithms are shown in Table 2 .
The comparison of the usage frequency and accuracy of different supervised learning algorithms are shown in Table 3 . It is observed that SVM has been used most frequently (29 out of 49 diseases that were predicted). This is followed by NB, which has been used in 23 articles. Although RF has been considered the second least number of times, it showed the highest percentage (i.e., 53%) in revealing the superior accuracy followed by SVM (i.e., 41%).
In Table 4 , the performance comparison of different supervised machine learning algorithms for most frequently modelled diseases is shown. It is observed that SVM showed the superior accuracy at most times for three diseases (e.g., heart disease, diabetes and Parkinson’s disease). For breast cancer, ANN showed the superior accuracy at most times.
A close investigation of Table 1 reveals an interesting result regarding the performance of different supervised learning algorithms. This result has also been reported in Table 4 . Consideration of only those articles that used clinical and demographic data (15 articles) reveals DT as to show the superior result at most times (6). Interestingly, SVM has been found the least time (1) to show the superior result although it showed the superior accuracy at most times for heart disease, diabetes and Parkinson’s disease (Table 4 ). In other 33 articles that used research data other than ‘clinical and demographic’ type, SVM and RF have been found to show the superior accuracy at most times (12) and second most times (7), respectively. In articles where 10-fold and 5-fold validation methods were used, SVM has been found to show the superior accuracy at most times (5 and 3 times, respectively). On the other side, articles where no method was used for validation, ANN has been found at most times to show the superior accuracy. Figure 12 further illustrates the superior performance of SVM. Performance statistics from Table 4 have been used in a normalised way to draw these two graphs. Fig. 12 a illustrates the ROC graph for the four diseases (i.e., Heart disease, Diabetes, Breast cancer and Parkinson’s disease) under the ‘ disease names that were modelled ’ criterion. The ROC graph based on the ‘ validation method followed ’ criterion has been presented in Fig. 12 b.
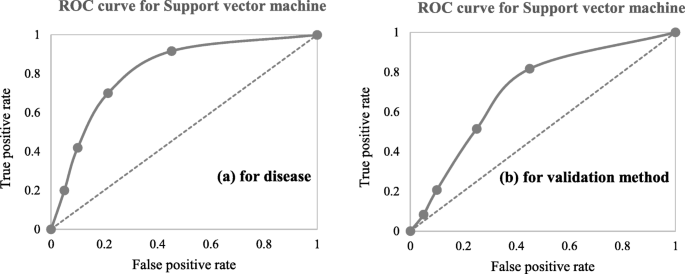
Illustration of the superior performance of the Support vector machine using ROC graphs (based on the data from Table 4 ) – ( a ) for disease names that were modelled; and ( b ) for validation methods that were followed
To avoid the risk of selection bias, from the literature we extracted those articles that used more than one supervised machine learning algorithm. The same supervised learning algorithm can generate different results across various study settings. There is a chance that a performance comparison between two supervised learning algorithms can generate imprecise results if they were employed in different studies separately. On the other side, the results of this study could suffer a variable selection bias from individual articles considered in this study. These articles used different variables or measures for disease prediction. We noticed that the authors of these articles did not consider all available variables from the corresponding research datasets. The inclusion of a new variable could improve the accuracy of an underperformed algorithm considered in the underlying study, and vice versa. This is one of the limitations of this study. Another limitation of this study is that we considered a broader level classification of supervised machine learning algorithms to make a comparison among them for disease prediction. We did not consider any sub-classifications or variants of any of the algorithms considered in this study. For example, we did not make any performance comparison between least-square and sparse SVMs; instead of considering them under the SVM algorithm. A third limitation of this study is that we did not consider the hyperparameters that were chosen in different articles of this study in comparing multiple supervised machine learning algorithms. It has been argued that the same machine learning algorithm can generate different accuracy results for the same data set with the selection of different values for the underlying hyperparameters [ 81 , 82 ]. The selection of different kernels for support vector machines can result a variation in accuracy outcomes for the same data set. Similarly, a random forest could generate different results, while splitting a node, with the changes in the number of decision trees within the underlying forest.
This research attempted to study comparative performances of different supervised machine learning algorithms in disease prediction. Since clinical data and research scope varies widely between disease prediction studies, a comparison was only possible when a common benchmark on the dataset and scope is established. Therefore, we only chose studies that implemented multiple machine learning methods on the same data and disease prediction for comparison. Regardless of the variations on frequency and performances, the results show the potential of these families of algorithms in the disease prediction.
Availability of data and materials
The data used in this study can be extracted from online databases. The detail of this extraction has been described within the manuscript.
Abbreviations
Area under the ROC curve
Decision Tree
False negative
False positive
Mean absolute error
Running mean square error
Receiver operating characteristic
True negative
True positive
T. M. Mitchell, “Machine learning WCB”: McGraw-Hill Boston, MA:, 1997.
Google Scholar
Sebastiani F. Machine learning in automated text categorization. ACM Comput Surveys (CSUR). 2002;34(1):1–47.
Sinclair C, Pierce L, Matzner S. An application of machine learning to network intrusion detection. In: Computer Security Applications Conference, 1999. (ACSAC’99) Proceedings. 15th Annual; 1999. p. 371–7. IEEE.
Sahami M, Dumais S, Heckerman D, Horvitz E. A Bayesian approach to filtering junk e-mail. In: Learning for Text Categorization: Papers from the 1998 workshop, vol. 62; 1998. p. 98–105. Madison, Wisconsin.
Aleskerov E, Freisleben B, Rao B. Cardwatch: A neural network based database mining system for credit card fraud detection. In: Computational Intelligence for Financial Engineering (CIFEr), 1997., Proceedings of the IEEE/IAFE 1997; 1997. p. 220–6. IEEE.
Kim E, Kim W, Lee Y. Combination of multiple classifiers for the customer's purchase behavior prediction. Decis Support Syst. 2003;34(2):167–75.
Mahadevan S, Theocharous G. “Optimizing Production Manufacturing Using Reinforcement Learning,” in FLAIRS Conference; 1998. p. 372–7.
Yao D, Yang J, Zhan X. A novel method for disease prediction: hybrid of random forest and multivariate adaptive regression splines. J Comput. 2013;8(1):170–7.
R. S. Michalski, J. G. Carbonell, and T. M. Mitchell, Machine learning: an artificial intelligence approach. Springer Science & Business Media, 2013.
Culler SD, Parchman ML, Przybylski M. Factors related to potentially preventable hospitalizations among the elderly. Med Care. 1998;1:804–17.
Uddin MS, Hossain L. Social networks enabled coordination model for cost Management of Patient Hospital Admissions. J Healthc Qual. 2011;33(5):37–48.
PubMed Google Scholar
Lee PP, et al. Cost of patients with primary open-angle glaucoma: a retrospective study of commercial insurance claims data. Ophthalmology. 2007;114(7):1241–7.
Davis DA, Chawla NV, Christakis NA, Barabási A-L. Time to CARE: a collaborative engine for practical disease prediction. Data Min Knowl Disc. 2010;20(3):388–415.
McCormick T, Rudin C, Madigan D. A hierarchical model for association rule mining of sequential events: an approach to automated medical symptom prediction; 2011.
Yiannakoulias N, Schopflocher D, Svenson L. Using administrative data to understand the geography of case ascertainment. Chron Dis Can. 2009;30(1):20–8.
CAS Google Scholar
Fisher ES, Malenka DJ, Wennberg JE, Roos NP. Technology assessment using insurance claims: example of prostatectomy. Int J Technol Assess Health Care. 1990;6(02):194–202.
CAS PubMed Google Scholar
Farran B, Channanath AM, Behbehani K, Thanaraj TA. Predictive models to assess risk of type 2 diabetes, hypertension and comorbidity: machine-learning algorithms and validation using national health data from Kuwait-a cohort study. BMJ Open. 2013;3(5):e002457.
PubMed PubMed Central Google Scholar
Ahmad LG, Eshlaghy A, Poorebrahimi A, Ebrahimi M, Razavi A. Using three machine learning techniques for predicting breast cancer recurrence. J Health Med Inform. 2013;4(124):3.
Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–9.
Demšar J. Statistical comparisons of classifiers over multiple data sets. J Mach Learn Res. 2006;7:1–30.
Palaniappan S, Awang R. Intelligent heart disease prediction system using data mining techniques. In: Computer Systems and Applications, 2008. AICCSA 2008. IEEE/ACS International Conference on; 2008. p. 108–15. IEEE.
Hosmer Jr DW, Lemeshow S, Sturdivant RX. Applied logistic regression. Wiley; 2013.
Joachims T. Making large-scale SVM learning practical. SFB 475: Komplexitätsreduktion Multivariaten Datenstrukturen, Univ. Dortmund, Dortmund, Tech. Rep. 1998. p. 28.
Quinlan JR. Induction of decision trees. Mach Learn. 1986;1(1):81–106.
Cruz JA, Wishart DS. Applications of machine learning in cancer prediction and prognosis. Cancer Informat. 2006;2:59–77.
Breiman L. Random forests. Mach Learn. 2001;45(1):5–32.
Lindley DV. Fiducial distributions and Bayes’ theorem. J Royal Stat Soc. Series B (Methodological). 1958;1:102–7.
I. Rish, “An empirical study of the naive Bayes classifier,” in IJCAI 2001 workshop on empirical methods in artificial intelligence, 2001, vol. 3, 22, pp. 41–46: IBM New York.
Cover T, Hart P. Nearest neighbor pattern classification. IEEE Trans Inf Theory. 1967;13(1):21–7.
McCulloch WS, Pitts W. A logical calculus of the ideas immanent in nervous activity. Bull Math Biophys. 1943;5(4):115–33.
Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature. 1986;323(6088):533.
Falagas ME, Pitsouni EI, Malietzis GA, Pappas G. Comparison of PubMed, Scopus, web of science, and Google scholar: strengths and weaknesses. FASEB J. 2008;22(2):338–42.
PubMed. (2018). https://www.ncbi.nlm.nih.gov/pubmed/ .
Kavakiotis I, Tsave O, Salifoglou A, Maglaveras N, Vlahavas I, Chouvarda I. Machine learning and data mining methods in diabetes research. Comput Struct Biotechnol J. 2017;15:104–16.
Pedregosa F, et al. Scikit-learn: Machine learning in Python. J Mach Learn Res. 2011;12:2825–30.
Borah MS, Bhuyan BP, Pathak MS, Bhattacharya P. Machine learning in predicting hemoglobin variants. Int J Mach Learn Comput. 2018;8(2):140–3.
Fawcett T. An introduction to ROC analysis. Pattern Recogn Lett. 2006;27(8):861–74.
Aneja S, Lal S. Effective asthma disease prediction using naive Bayes—Neural network fusion technique. In: International Conference on Parallel, Distributed and Grid Computing (PDGC); 2014. p. 137–40. IEEE.
Ayer T, Chhatwal J, Alagoz O, Kahn CE Jr, Woods RW, Burnside ES. Comparison of logistic regression and artificial neural network models in breast cancer risk estimation. Radiographics. 2010;30(1):13–22.
Lundin M, Lundin J, Burke H, Toikkanen S, Pylkkänen L, Joensuu H. Artificial neural networks applied to survival prediction in breast cancer. Oncology. 1999;57(4):281–6.
Delen D, Walker G, Kadam A. Predicting breast cancer survivability: a comparison of three data mining methods. Artif Intell Med. 2005;34(2):113–27.
Chen M, Hao Y, Hwang K, Wang L, Wang L. Disease prediction by machine learning over big data from healthcare communities. IEEE Access. 2017;5:8869–79.
Cai L, Wu H, Li D, Zhou K, Zou F. Type 2 diabetes biomarkers of human gut microbiota selected via iterative sure independent screening method. PLoS One. 2015;10(10):e0140827.
Malik S, Khadgawat R, Anand S, Gupta S. Non-invasive detection of fasting blood glucose level via electrochemical measurement of saliva. SpringerPlus. 2016;5(1):701.
Mani S, Chen Y, Elasy T, Clayton W, Denny J. Type 2 diabetes risk forecasting from EMR data using machine learning. In: AMIA annual symposium proceedings, vol. 2012; 2012. p. 606. American Medical Informatics Association.
Tapak L, Mahjub H, Hamidi O, Poorolajal J. Real-data comparison of data mining methods in prediction of diabetes in Iran. Healthc Inform Res. 2013;19(3):177–85.
Sisodia D, Sisodia DS. Prediction of diabetes using classification algorithms. Procedia Comput Sci. 2018;132:1578–85.
Yang J, Yao D, Zhan X, Zhan X. Predicting disease risks using feature selection based on random forest and support vector machine. In: International Symposium on Bioinformatics Research and Applications; 2014. p. 1–11. Springer.
Juhola M, Joutsijoki H, Penttinen K, Aalto-Setälä K. Detection of genetic cardiac diseases by Ca 2+ transient profiles using machine learning methods. Sci Rep. 2018;8(1):9355.
Long NC, Meesad P, Unger H. A highly accurate firefly based algorithm for heart disease prediction. Expert Syst Appl. 2015;42(21):8221–31.
Jin B, Che C, Liu Z, Zhang S, Yin X, Wei X. Predicting the risk of heart failure with ehr sequential data modeling. IEEE Access. 2018;6:9256–61.
Puyalnithi T, Viswanatham VM. Preliminary cardiac disease risk prediction based on medical and behavioural data set using supervised machine learning techniques. Indian J Sci Technol. 2016;9(31):1–5.
Forssen H, et al. Evaluation of Machine Learning Methods to Predict Coronary Artery Disease Using Metabolomic Data. Stud Health Technol Inform. 2017;235: IOS Press:111–5.
Tang Z-H, Liu J, Zeng F, Li Z, Yu X, Zhou L. Comparison of prediction model for cardiovascular autonomic dysfunction using artificial neural network and logistic regression analysis. PLoS One. 2013;8(8):e70571.
CAS PubMed PubMed Central Google Scholar
Toshniwal D, Goel B, Sharma H. Multistage Classification for Cardiovascular Disease Risk Prediction. In: International Conference on Big Data Analytics; 2015. p. 258–66. Springer.
Alonso DH, Wernick MN, Yang Y, Germano G, Berman DS, Slomka P. Prediction of cardiac death after adenosine myocardial perfusion SPECT based on machine learning. J Nucl Cardiol. 2018;1:1–9.
Mustaqeem A, Anwar SM, Majid M, Khan AR. Wrapper method for feature selection to classify cardiac arrhythmia. In: Engineering in Medicine and Biology Society (EMBC), 39th Annual International Conference of the IEEE; 2017. p. 3656–9. IEEE.
Mansoor H, Elgendy IY, Segal R, Bavry AA, Bian J. Risk prediction model for in-hospital mortality in women with ST-elevation myocardial infarction: a machine learning approach. Heart Lung. 2017;46(6):405–11.
Kim J, Lee J, Lee Y. Data-mining-based coronary heart disease risk prediction model using fuzzy logic and decision tree. Healthc Inform Res. 2015;21(3):167–74.
Taslimitehrani V, Dong G, Pereira NL, Panahiazar M, Pathak J. Developing EHR-driven heart failure risk prediction models using CPXR (log) with the probabilistic loss function. J Biomed Inform. 2016;60:260–9.
Anbarasi M, Anupriya E, Iyengar N. Enhanced prediction of heart disease with feature subset selection using genetic algorithm. Int J Eng Sci Technol. 2010;2(10):5370–6.
Bhatla N, Jyoti K. An analysis of heart disease prediction using different data mining techniques. Int J Eng. 2012;1(8):1–4.
Thenmozhi K, Deepika P. Heart disease prediction using classification with different decision tree techniques. Int J Eng Res Gen Sci. 2014;2(6):6–11.
Tamilarasi R, Porkodi DR. A study and analysis of disease prediction techniques in data mining for healthcare. Int J Emerg Res Manag Technoly ISSN. 2015;1:2278–9359.
Marikani T, Shyamala K. Prediction of heart disease using supervised learning algorithms. Int J Comput Appl. 2017;165(5):41–4.
Lu P, et al. Research on improved depth belief network-based prediction of cardiovascular diseases. J Healthc Eng. 2018;2018:1–9.
Khateeb N, Usman M. Efficient Heart Disease Prediction System using K-Nearest Neighbor Classification Technique. In: Proceedings of the International Conference on Big Data and Internet of Thing; 2017. p. 21–6. ACM.
Patel SB, Yadav PK, Shukla DD. Predict the diagnosis of heart disease patients using classification mining techniques. IOSR J Agri Vet Sci (IOSR-JAVS). 2013;4(2):61–4.
Venkatalakshmi B, Shivsankar M. Heart disease diagnosis using predictive data mining. Int J Innovative Res Sci Eng Technol. 2014;3(3):1873–7.
Ani R, Sasi G, Sankar UR, Deepa O. Decision support system for diagnosis and prediction of chronic renal failure using random subspace classification. In: Advances in Computing, Communications and Informatics (ICACCI), 2016 International Conference on; 2016. p. 1287–92. IEEE.
Islam MM, Wu CC, Poly TN, Yang HC, Li YC. Applications of Machine Learning in Fatty Live Disease Prediction. In: 40th Medical Informatics in Europe Conference, MIE 2018; 2018. p. 166–70. IOS Press.
Lynch CM, et al. Prediction of lung cancer patient survival via supervised machine learning classification techniques. Int J Med Inform. 2017;108:1–8.
Chen C-Y, Su C-H, Chung I-F, Pal NR. Prediction of mammalian microRNA binding sites using random forests. In: System Science and Engineering (ICSSE), 2012 International Conference on; 2012. p. 91–5. IEEE.
Eskidere Ö, Ertaş F, Hanilçi C. A comparison of regression methods for remote tracking of Parkinson’s disease progression. Expert Syst Appl. 2012;39(5):5523–8.
Chen H-L, et al. An efficient diagnosis system for detection of Parkinson’s disease using fuzzy k-nearest neighbor approach. Expert Syst Appl. 2013;40(1):263–71.
Behroozi M, Sami A. A multiple-classifier framework for Parkinson’s disease detection based on various vocal tests. Int J Telemed Appl. 2016;2016:1–9.
Hussain L, et al. Prostate cancer detection using machine learning techniques by employing combination of features extracting strategies. Cancer Biomarkers. 2018;21(2):393–413.
Zupan B, DemšAr J, Kattan MW, Beck JR, Bratko I. Machine learning for survival analysis: a case study on recurrence of prostate cancer. Artif Intell Med. 2000;20(1):59–75.
Hung C-Y, Chen W-C, Lai P-T, Lin C-H, Lee C-C. Comparing deep neural network and other machine learning algorithms for stroke prediction in a large-scale population-based electronic medical claims database. In: Engineering in Medicine and Biology Society (EMBC), 2017 39th Annual International Conference of the IEEE, vol. 1; 2017. p. 3110–3. IEEE.
Atlas L, et al. A performance comparison of trained multilayer perceptrons and trained classification trees. Proc IEEE. 1990;78(10):1614–9.
Lucic M, Kurach K, Michalski M, Bousquet O, Gelly S. Are GANs created equal? a large-scale study. In: Proceedings of the 32nd International Conference on Neural Information Processing Systems; 2018. p. 698–707. Curran Associates Inc.
Levy O, Goldberg Y, Dagan I. Improving distributional similarity with lessons learned from word embeddings. Trans Assoc Comput Linguistics. 2015;3:211–25.
Download references
Acknowledgements
Not applicable.
This study did not receive any funding.
Author information
Authors and affiliations.
Complex Systems Research Group, Faculty of Engineering, The University of Sydney, Room 524, SIT Building (J12), Darlington, NSW, 2008, Australia
Shahadat Uddin, Arif Khan & Md Ekramul Hossain
Health Market Quality Research Stream, Capital Markets CRC, Level 3, 55 Harrington Street, Sydney, NSW, Australia
Faculty of Medicine and Health, School of Medical Sciences, The University of Sydney, Camperdown, NSW, 2006, Australia
Mohammad Ali Moni
You can also search for this author in PubMed Google Scholar
Contributions
SU: Originator of the idea, data analysis and writing. AK: Data analysis and writing. MEH: Data analysis and writing. MAM: Data analysis and critical review of the manuscript. All authors have read and approved the manuscript.
Corresponding author
Correspondence to Shahadat Uddin .
Ethics declarations
Ethics approval and consent to participate, consent for publication, competing interests.
The authors declare that they do not have any competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License ( http://creativecommons.org/licenses/by/4.0/ ), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated.
Reprints and permissions
About this article
Cite this article.
Uddin, S., Khan, A., Hossain, M. et al. Comparing different supervised machine learning algorithms for disease prediction. BMC Med Inform Decis Mak 19 , 281 (2019). https://doi.org/10.1186/s12911-019-1004-8
Download citation
Received : 28 January 2019
Accepted : 11 December 2019
Published : 21 December 2019
DOI : https://doi.org/10.1186/s12911-019-1004-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Machine learning
- Medical data
- Disease prediction
BMC Medical Informatics and Decision Making
ISSN: 1472-6947
- General enquiries: [email protected]
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 24 February 2021
Infectious disease outbreak prediction using media articles with machine learning models
- Juhyeon Kim 1 , 2 &
- Insung Ahn 1 , 2
Scientific Reports volume 11 , Article number: 4413 ( 2021 ) Cite this article
8917 Accesses
13 Citations
Metrics details
- Computer science
- Epidemiology
- Infectious diseases
- Information technology
When a newly emerging infectious disease breaks out in a country, it brings critical damage to both human health conditions and the national economy. For this reason, apprehending which disease will newly emerge, and preparing countermeasures for that disease, are required. Many different types of infectious diseases are emerging and threatening global human health conditions. For this reason, the detection of emerging infectious disease pattern is critical. However, as the epidemic spread of infectious disease occurs sporadically and rapidly, it is not easy to predict whether an infectious disease will emerge or not. Furthermore, accumulating data related to a specific infectious disease is not easy. For these reasons, finding useful data and building a prediction model with these data is required. The Internet press releases numerous articles every day that rapidly reflect currently pending issues. Thus, in this research, we accumulated Internet articles from Medisys that were related to infectious disease, to see if news data could be used to predict infectious disease outbreak. Articles related to infectious disease from January to December 2019 were collected. In this study, we evaluated if newly emerging infectious diseases could be detected using the news article data. Support Vector Machine (SVM), Semi-supervised Learning (SSL), and Deep Neural Network (DNN) were used for prediction to examine the use of information embedded in the web articles: and to detect the pattern of emerging infectious disease.
Similar content being viewed by others
Crowdsourcing and machine learning approaches for extracting entities indicating potential foodborne outbreaks from social media
Dandan Tao, Dongyu Zhang, … Hao Feng
Forecasting virus outbreaks with social media data via neural ordinary differential equations
Matías Núñez, Nadia L. Barreiro, … Christopher Rackauckas
Development of an early alert model for pandemic situations in Germany
Danqi Wang, Manuel Lentzen, … Holger Fröhlich
Introduction
The spread of middle East respiratory syndrome (MERS) in 2015 caused 185 confirmed cases and 36 deaths 1 . The first outbreak of MERS in the Republic of Korea (Korea) occurred on May 2015, after a 68-year-old man returned from a business trip to several Middle East countries. As Korea could not predict if MERS might flow across the border, MERS not only threatened public health, but also caused huge economic loss in many different categories, including the tourist industry and social activity. Such a situation indicates that judging if an infectious disease will influx from other countries or not in advance is an important issue to minimize the damage that ensues. MERS was first reported in September 2012 from Saudi Arabia, and was reported from several European countries, before MERS occurred in Korea during 2015 1 . As MERS was not a commonly known disease in Korea, there was indifference to it before it occurred. However, if it was possible to predict that MERS might flow into Korea while it was spreading around the world, Korea could have prepared for the outbreak of the MERS to minimize the damage it caused. On the other hand, while MERS was spreading through several continents, Ebola spread through 5 different countries in Western Africa, infecting more than 6,500 people, and killing more than 3000 people 2 . Even though Ebola outbreaks occurred a few times on the Africa continent, the 2014 pandemic was the biggest one 3 . The 2014 Ebola pandemic in Western Africa showed a fatality rate of over 50%. However unlike MERS, Ebola, did not spread throughout other continents.
Many different infectious diseases threaten lives worldwide. Some diseases, like MERS, cause pandemics, spreading from country to country over continents, while some do not spread over continents, but like Ebola, circulate only in a few countries. As infectious disease issues arise worldwide, many researches were conducted to estimate and predict the occurrence of infectious diseases. Authors of 4 , 5 developed infectious disease spread simulation models using mathematical models. These research efforts utilized susceptible infected recovered (SIR) models to build an infectious disease spread simulation model, and suggest strategies to control infectious disease and maximize the effect of vaccination with the results from the simulation models. Commonly, these SIR simulation models concern the population of the area the model is based on and the characteristic of the disease, such as infection rate, incubation rate, and recovery rate. Some research considers the passengers of flights crossing borders to explain how infectious disease spreads abroad 6 . Moreover, authors of 7 claimed that infectious disease epidemics can be related to climate and climatic events, such as El Nino. According to the existing research reports above, the occurrence of infectious diseases varies depending on many different reasons, such as climate, lifestyle of countries, diplomatic relations between countries, or population. Thus, it is important to collect and use the latest data for future infectious disease outbreak prediction. However, the degree of these features for each country varies according to the passage of time. For example, El Nino changes the climatic attributes throughout the world, digitalization changes the lifestyle of human, and the number of travelers or the amount of trade between countries may change dramatically for political reasons. Consequently, constructing an infectious disease outbreak prediction model considering all these features is a challenging matter. However, as infectious disease spreads based on all these features, it may be possible to assert that the rate of particular infectious disease occurrence in a particular country connotes the information mentioned above. This means that we can assume that some infectious disease occurs in a particular country, because the conditions of certain features, such as climate, population, lifestyle, and the number of incoming travelers exceed thresholds for the disease in that country. With this assumption, it is possible to forecast if an infectious disease that has not occurred recently in a particular country will break out or not in that country, by analyzing the patterns of many different types of infectious diseases occurring in different countries.
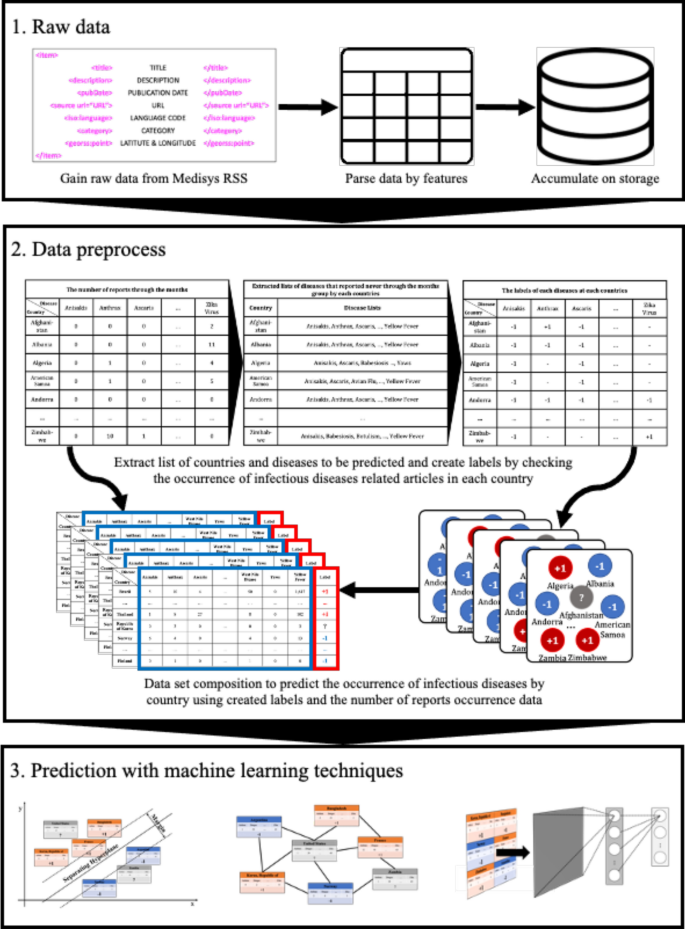
Normally, when an infectious disease breaks out, the press media publish articles concerning the disease. When the seriousness of the disease becomes higher for some reason, like the increase in the number of infected people, the number of published articles also increases. In other words, the number of articles reported related to a particular disease in a particular country reflects how severe the disease is in that country. Furthermore, media articles and reports are updated in real-time through the Internet service worldwide, which offers the advantage of accumulating the latest data immediately, while collecting actual surveillance data of numerous disease types from countries worldwide is a difficult task 8 . Therefore, various attempts have been made to utilize media article data to predict an epidemic outbreak. Most of these studies, utilizing data from online media articles, try to figure out the epidemics occurring in specific country. In study 9 , media articles related to specific infectious diseases that occurred in the United States, China, and India respectively were collected, and based on this, the temporal topic trend was compared with the actual disease case count. The outbreak of whooping cough, rabies, salmonellosis, and E. coli infection in the United States, H7N9, hand, foot, and mouth disease, and dengue in China, and acute diarrheal disease, dengue, and malaria in India were estimated by proposing method. This allowed the authors to successfully capture the dynamics of disease outbreak by the temporal topic trends obtained through media articles. In other words, the degree of the temporal topic trend for a specific infectious disease in such a specific country can actually indicate the severity of the infectious disease in that country. Furthermore, in a study proposing a method to monitor infectious diseases using online news media data, the proposed model was applied to the outbreak of dengue fever in India and the outbreak of zika virus in Brazil 10 . In the study, using the collected international newspaper data and local newspaper data, the number of news reports related to each disease was calculated, and how similar the number of actual disease cases was. The authors of the study argue that there is a possibility to build a surveillance system using news data even in developing countries that do not have a surveillance systems yet. The authors of 8 , 11 suggested a method of predicting the occurrence of infectious diseases by extracting keywords with high relevance to specific infectious diseases instead of the simple number of occurrences of media articles related to a specific infectious disease. All of the aforementioned studies suggest a method to estimate the number of patients with a specific infectious disease in a specific country using online media article data. The existing studies showed the potential that online media article data can make a great contribution to the prediction of infectious diseases. However, since previous studies have focused on establishing an outbreak surveillance system, such as measuring the number of existing infectious diseases in a specific region, there is a limitation that only a limited number of infectious diseases can be treated in a limited number of countries. In other words, it cannot handle various kinds of infectious diseases occurring in various countries because the country and the type of infectious disease are specified. In addition, the previous infectious disease prediction studies have successfully established a surveillance system for infectious diseases that have seasonality or have been present in certain countries, but there is a limitation that it is impossible to predict the occurrence of infectious diseases that have not occurred. Thus, this study proposes a methodology for predicting the occurrence of various infectious diseases that did not occur for 6 months in various countries around the world by analyzing media article data. The remainder of this paper is organized as follows. “ Methods ” section explains which data is used for infectious disease outbreak prediction, and introduces the three machine learning models, semi-supervised learning (SSL), support vector machine (SVM), and deep neural network (DNN). “ Experiments ” section details the performance measures, and the experimental settings and results. Finally, “ Results ” and “ Conclusion ” sections present our discussions and conclusions, respectively.
Nowadays, as the Internet service is supplied worldwide, people obtain information using the Internet service easily and rapidly. Even news articles are being published through the Internet, unlike in the past, when they were printed on paper and delivered. Accordingly, articles and reports related to infectious diseases are also being published and updated through the Internet media in real-time. In other words, unlike in the past, the Internet media has made it easy to obtain information about the seriousness of infectious disease issues around the world today. Thus, in this research, we collected articles and reports related to 115 different infectious diseases from Medisys, to predict if a particular infectious disease that had not occurred for several months in a particular country will break out in that country. Medisys serves news articles and reports of infectious disease published worldwide every day in real-time 12 . Articles and reports provided by Medisys are classified by disease, and include the date and time they were published, and the latitude and longitude of the information where the outbreak of disease occurred. Every articles are also published in rich site summary (RSS) form. RSS is a method of displaying content primarily used on news or blog sites. If website administrators display website content in RSS format, recipients of this information may use it in different formats. Figure 1 shows examples of data provided by Medisys in RSS form and their components. The information of each article is displayed between < item > and < /item > , and data such as article title, description, publication date, original url, language code, category indicating the name of the disease, latitude and longitude are displayed. Even though the Medisys reports do not provide where the articles are published, it is possible to track where they were published by analyzing the latitude and longitude information. We accumulated data from Medisys for January to December 2019. This data consisted of 115,279 articles published in 237 different countries. As described in Fig. 2 , the number of articles per nation, and infectious disease were extracted from the data, and utilized in this study. However, some poor and developing countries, especially if involved in wars, have less opportunity to publish digital data. Furthermore, the population sizes by country also varies which may affect the number of published articles. For these reasons, data is normalized between 0 and 1 by each country to adjust values measured on different scales. Figure 3 shows the reorganized data.
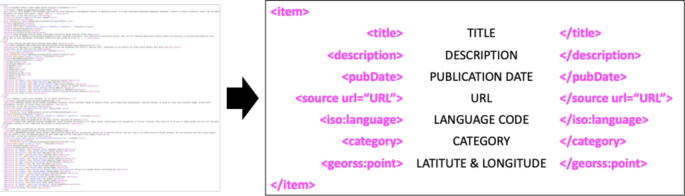
Examples of data provided by Medisys in the form of RSS and components of RSS provided by Medisys.
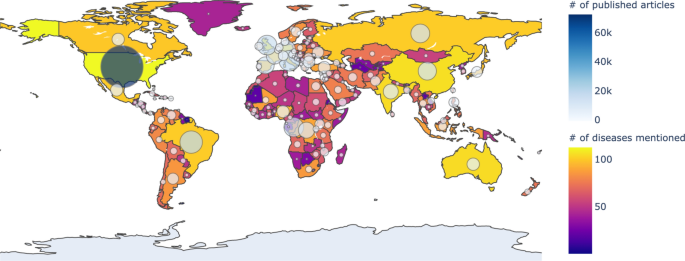
The number of articles published in each country collected from Medisys from January to December 2019: the closer the color of the country to yellow, the more diseases occurred, and the larger the circle, the more articles have occurred. The figure was created in Python3 using the Basemap Toolkit.
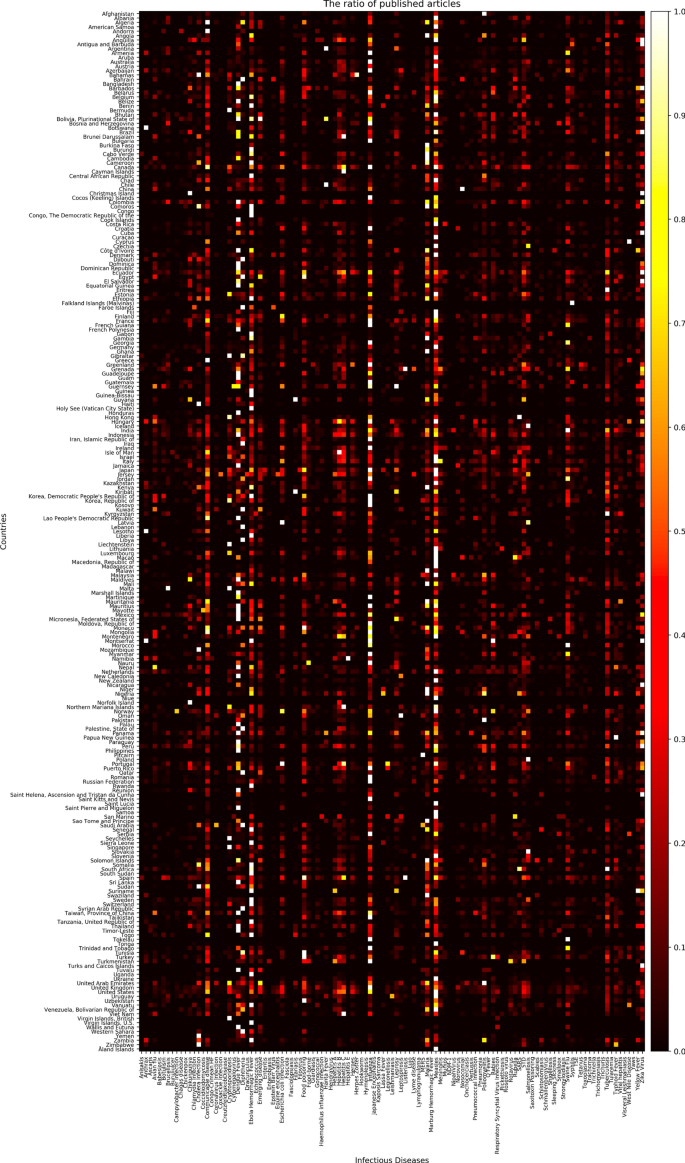
The number of articles related to each disease by country collected from Medisys for 2019 from January to December (data is normalized, thus the brighter the color, the more articles; the darker, the less articles).
To apply the constructed data to machine learning models to predict if disease that had not occurred for several months in a particular country would occur or not, the data set was preprocessed as follows. For example, as shown in Fig. 4 , Table A extracted the number of articles related to 115 different diseases by 237 countries during the 6 month period February to July 2019. From Table A, a disease list that contains ‘0′, which means diseases never occurred from each country, was extracted and listed in Table B by country. Each disease listed in Table B is considered, as it may have the potential for outbreak, because it has not yet occurred in each country. Table C is the data from August to October 2019, 3 months after July 2019. Then if the data of a particular disease for a particular country is 0 in both Tables A and C, the label of the disease of the country becomes ‘ − 1′; while when Table A is 0, but Table C is > 0, the label becomes ‘ + 1′. These labels can be arranged as in Table D.
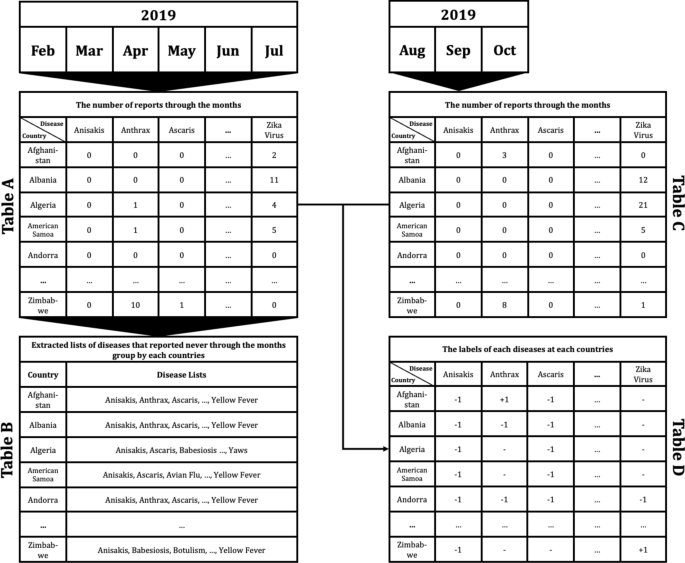
Example of data preprocessing to predict infectious disease outbreak for 3 months after July 2019, using report count data from February to July 2019: Table A indicates the number of reports concerning each disease in each country from February to July 2019. Table B shows the lists of diseases that reported none during the 6 month period February to July 2019 in each country. Table C shows the number of counted reports related to listed diseases in each country from August to October in 2019. Finally, Table D shows the labels of each disease for each country. ‘ + 1′ indicates that the disease occurred in the country between August and October; in contrast, ‘ − 1′ indicates that the disease did not occur, while ‘ − ’ means that the disease had already occurred during the period February to July, thus the disease for the country does not display a label.
Once the data has been preprocessed as shown in Fig. 4 , it is possible to select a list of what infectious diseases should be predicted in each country, as shown in Table B, and based on this list, it is possible to create a label set for each country and for each disease, as shown in Table D. With the preprocessed data, the data set for the prediction models of each disease by country can be organized as shown in Fig. 5 . Every node in Fig. 5 is composed of data in Table A in Fig. 4 . In Fig. 5 , if more than a single report related to the infectious disease by country occurred, then ‘ + 1′ is labeled, and in contrast, in the case of a report that was not reported in Table A of Fig. 4 , ‘ − 1′ is labeled ,while unlabeled nodes ‘?’ are listed in Table B of Fig. 4 . In other words, each square shown in Fig. 5 is a set of labels for predicting disease outbreak in each country, and the overall data structure of each square can be represented as shown in Fig. 6 . In Fig. 6 , the number in each column is the number of media articles related to each infectious disease in each country that occurred during the specified period. For all unlabeled data, a data set as shown in Fig. 6 is formed based on label of Fig. 5 , and each data set is applied to machine learning models to predict the occurrence of a specific infectious disease in a specific country.
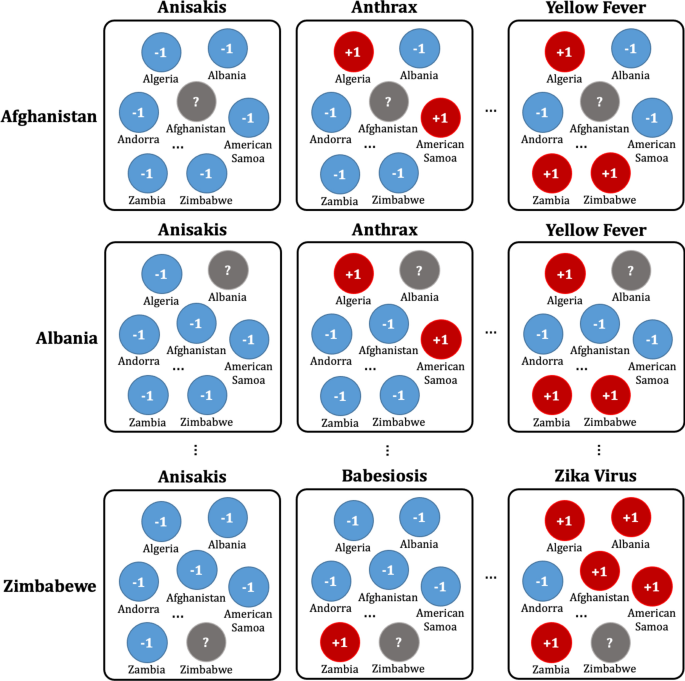
Data set composition for the prediction model for each disease by country: For example, in the first row, the data set of diseases for Afghanistan is listed in the first row of Table B of Fig. 4 . Nodes with ‘ + 1′ indicate that the reports related to the disease occurred more than once in the country, while nodes with ‘ − 1′ indicate that the reports related to the diseases never occurred in Table A of Fig. 4 .
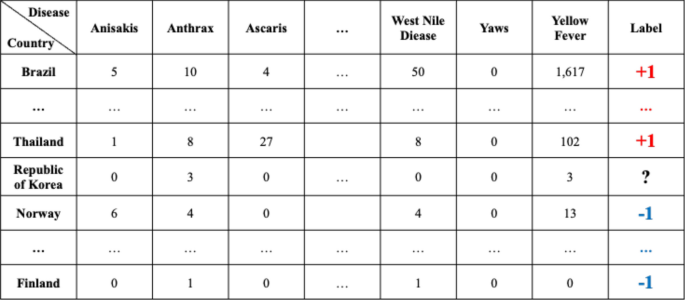
The overall data structure of each square shown in Fig. 5 .
In this research, we adapted three different machine learning models to investigate if early disease outbreak detection would be possible using media articles and reports related to infectious disease, and compared the performance of the models. Three representative models, that is, support vector machine (SVM), which shows good performance consistently through various fields; semi-supervised learning (SSL), which shows good performance when label imbalanced data sets are used; and deep neural network (DNN), which is a trending method showing outstanding performance, were used to perform prediction for disease occurrence. The model parameters of SVM, SSL, and DNN were searched over the following ranges. For SVM, the best prediction performances were identified from the combinations of { γ, C} ∈ {0.0001, 0.001, 0.01, 0.1, 1, 10} × {0.2, 0.4, 0.6, 0.8, 1} 13 . For SSL, k, which is a parameter to decide the number of neighbors was identified from k = {3, 7, 15, 20, 30}, and μ, which is a trade-off parameter, was identified from μ = {0.0001, 0.01, 1, 100, 1000}. Finally, DNN model was organized with 3 layers with batch size of 20 for each step. Dropouts are set as 0.3 for each layer, and Adam gradient descent optimization was applied, while epoch was set as 500. After disease outbreak prediction is made with each model, the model performance is calculated using Table D of Fig. 4 by comparing the prediction results with the corresponding infectious disease 3 months after the last date used as the training data. The order of progress from data preprocessing to prediction can be summarized as shown in Fig. 7 .
The order of progress from data preprocessing to prediction.
Ethics approval and consent to participate
This study did not involve human participants, data, or tissue. Institutional review board approval was not required.
Experiments
Media articles and reports that are published from January to December 2019 crawled from Medisys are used in this research. The crawled data includes the title of articles, description, published date and time, disease related to, and the latitude and longitude information. Parsing the data, counts of the number of daily articles related to each disease by country are extracted, and organized as a numerical dataset. A total of 115 different diseases and 237 different countries are concerned with the extracted dataset, and the average count of the number of daily articles is about 1300. Each data point is normalized between 0 and 1. As shown in Fig. 8 , experiments are done with two different strategies, setting the length of training data as (6 and 3) months, and the validation data as 3 months, respectively. It is discovered whether each model can predict whether diseases will break out or not by country during the 3 months after the training data of July to September, August to October, September to November, and October to December, respectively.
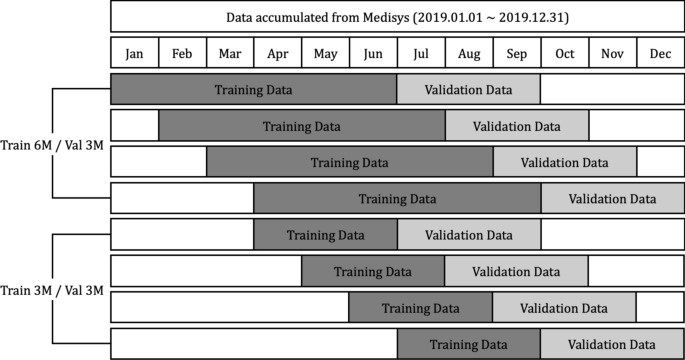
Using data crawled for a year, experiments are set as first, each model being trained using 6 months’ data, and predicting if the disease will outbreak or not; and second, each model being trained using 3 months’ data, and predicting if the disease will outbreak or not.
To measure the performance of each prediction model, AUC, Accuracy, and F1 score are used 14 , 15 . The AUC assesses the overall value of a classifier, which is a threshold-independent measure of model performance based on the receiver operating characteristic curve that plots the trade-offs between sensitivity and 1—specificity for all possible values of threshold. Accuracy is a measure of the total number of correct predictions when the value of the classification threshold is set to 0. Lastly, the F1 score can be interpreted as the weighted average of the precision and recall, where an F1 score reaches its best value at 1, while the worst score is 0.
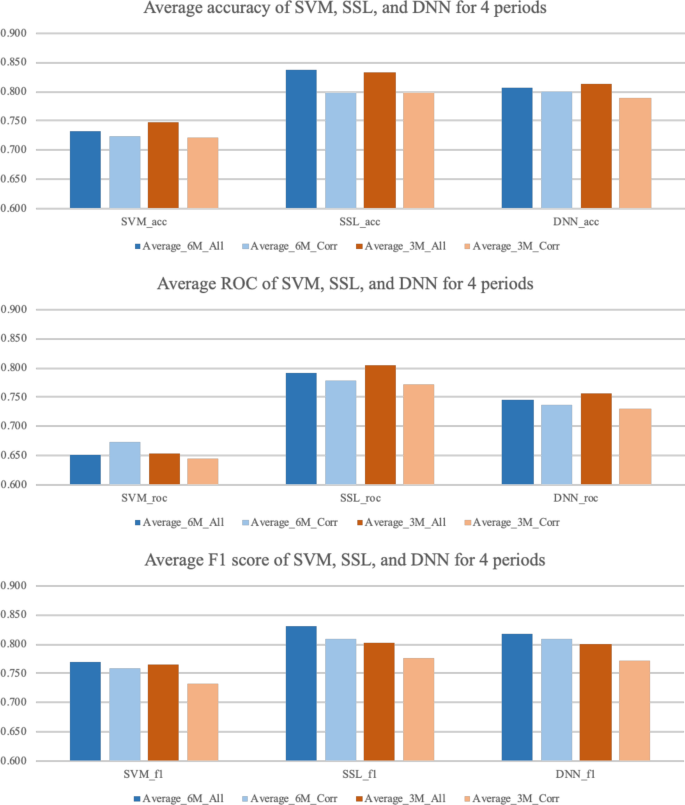
The results of the experiment are based on the expected accuracy of whether the diseases that had not been reported for (6 or 3) months will break out or not by country. Tables 1 and 2 show a comparison of the results with SVM, SSL, and DNN in terms of the accuracy, ROC, and F1 score. For each of the three models, the best performance was selected by searching over the respective model-parameter space. For each dataset, the best performance among the three models is marked in bold face. In terms of the accuracy, SSL shows the best performance, with an average accuracy of (0.838 and 0.834). In terms of the ROC, SSL delivers outstanding performance, with an average ROC of (0.791 and 0.805). Lastly, even in the F1 score case, SSL produces an average (0.832 and 0.802), which is the best of the three models. Figure 9 summarizes the performance of the three models in bar graphs. Even though SSL shows outstanding performance compared to other two models, SVM and DNN also show reasonable performance, showing average accuracy over 0.7, and F1 score over 0.75.

Accuracy, ROC, and F1 score of each validation data set period by each model, respectively.
In Fig. 10 , the prediction accuracy of SSL for 8 different experiments are shown in the world map. Some countries are not colored in the map because every kinds of diseases were mentioned through media articles in these countries. In other words, these countries had no diseases to be predicted. While prediction accuracy of most countries is over 0.8, there are some countries showing very low prediction accuracy. This is because countries showing low accuracy contains only small number of diseases to be predicted. Thus, a wrong prediction of any one would significantly reduce the accuracy of the prediction.
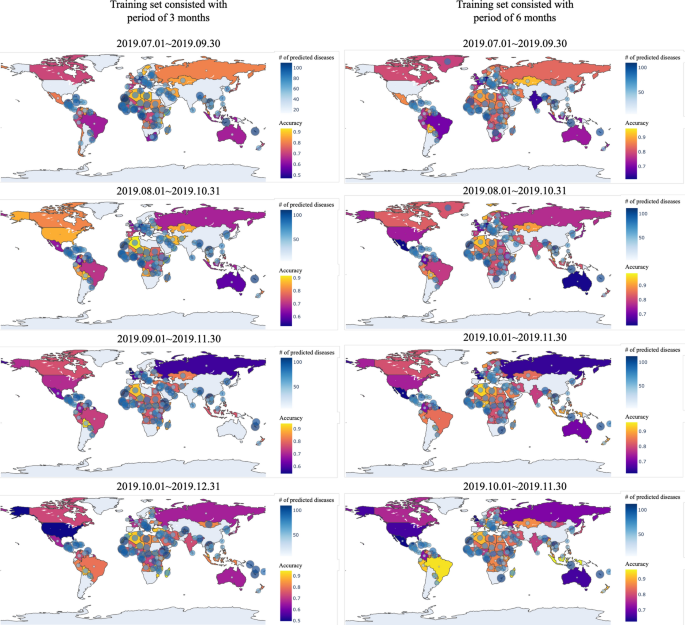
Prediction accuracy of SSL by each country: blue circles in the map indicate the number of predicted diseases for the country, and the closer the yellow, the more accurate the blue, the lower the accuracy. The figure was created in Python3 using the Basemap Toolkit.
In this research, the potential of utilizing media data to predict if an infectious disease will break out or not in a particular country using three of the most widely used machine learning models showed reasonable prediction performances. The occurrence of infectious diseases varies depending on many different reasons, such as climate, lifestyle of countries, diplomatic relations between countries, or population and so on. Therefore, similar types of infectious diseases are likely to occur in countries with similar comprehensive environments. In other words, countries with similar severity of various types of infectious diseases can be regarded as countries with similar environments. Thus, countries with similar infectious disease outbreak patterns can be identified by analyzing the patterns of severity of various types of infectious diseases between countries. Moreover, various existing studies have shown that the degree of incidence of media articles related to a specific infectious disease occurring in a specific country may indicate the severity of the disease in that country. Thus, this study attempted to predict the occurrence of specific infectious diseases in a specific country by analyzing the outbreak patterns of media articles related to various infectious diseases between countries. As the suggested method uses only media articles, even developing countries that have not yet constructed any disease surveillance systems, are able to forecast if particular infections will occur or not, because there are no critical limitations to accumulating such media articles.
Despite these advantages, further studies should be carried out in the near future to resolve several obstacles. First of all, the periods of training data and validation data were set by dividing the period of the year by the fourth quarter or half of the year from the data used for prediction, but a more systematic data time-setting strategy is needed, such as considering seasonal infectious diseases. Moreover, as Medisys does not provide old posts data, only about a year’s worth of data has been accumulated now since we started to collect Medisys data from the end of 2018. Therefore, when more data is collected, it is necessary to make predictions by later gathering additional data, and setting the duration of the training data to at least 1 year.
Second, even though all three models showed reasonable performance, it is necessary to discover methods to improve the performance of the prediction models. In this study, we also looked at whether it would be possible to improve the performance of prediction models when models are trained with data consisting of countries that show similar infectious disease occurrence patterns. Therefore, the prediction models for each country are trained using only data from countries with a correlation coefficient of 0.6 or higher, and Fig. 11 shows the performance of each prediction model. It was expected that the prediction results using data would consist of countries having disease outbreak pattern correlation coefficients of over 0.6; however, generally they showed worse performance. From this result, it can be inferred that even from countries where infectious disease patterns are dissimilar, the prediction model extracts useful information, and trains them. Thus, in further works, instead of feature selection, accumulating useful data, such as global air passenger data, which can represent the degree of relation between countries, is required to utilize them as weight for prediction models.
Prediction performance comparison between models using all countries and models using countries having a disease occurrence pattern correlation coefficient of over 0.6.
The biggest reasons why it is not easy to predict the exact incidence of infectious disease is that a variety of characteristics, such as the nature of the infectious disease, the geographical characteristics of where the infectious diseases occur, the characteristics of people living in the country, the way people live, the kinds of things that spread infectious diseases, and the degree of exchanges between countries should all be taken into account. Furthermore, as time goes by, the weather changes due to global warming, digitalization changes people’s lifestyles, and for many reasons, the status of countries that trade frequently with each other changes. For these reasons, it is challenging work to create a predictive model that takes all of these characteristics into account. However, as the pattern of infectious diseases varies from country to country due to these various reasons, infectious disease incidence data by country can be considered to contain this information. Therefore, in this research, we tried to predict which disease will occur or not in particular countries, analyzing media data accumulated from Medisys using several machine learning models. Our suggested method showed reasonable prediction performance by the three different trending machine learning models SVM, SSL, and DNN. It is thought that the proposed model could be used to prepare for the future outbreak of infectious diseases in various countries, including developing countries that lack proper disease surveillance systems.
Data availability
The datasets used during the current study are available from the corresponding author on reasonable request.
Abbreviations
Support vector machine
Semi-supervised learning
Deep neural network
Middle east respiratory syndrome
Susceptible infected recovered
Receiver operating characteristic
European Centre for Disease Prevention and Control. Middle East Respiratory Syndrome Coronavirus (MERSCoV). 21st Update (ECDC, Stockholm, 2015).
Google Scholar
Centers for Disease Control and Prevention. 2014 Ebola outbreak in West Africa: case counts, 2015. http://www.cdc.gov/vhf/ebola/outbreaks/2014-west-africa/case-counts.html . Accessed 6 April 2015.
Dixon, M. G. & Schafer, I. J. Ebola viral disease outbreak—West Africa, 2014. Morb. Mortal. Wkly Rep. 63 , 548–551 (2014).
Meyers, L. A. Contact network epidemiology: bond percolation applied to infectious disease prediction and control. Bull. Am. Math. Soc. 44 , 63–86 (2007).
Article MathSciNet Google Scholar
Dimitrov, N. B. & Meyerss, L. A. Mathematical approaches to infectious disease prediction and control. INFORMS Tutor. Oper. Res. 7 , 1–25 (2010).
Hufnagel, L., Brockmann, D. & Geisel, T. Forecast and control of epidemics in a globalized world. Proc. Natl. Acad. Sci. U.S.A. 101 , 15124–15129 (2004).
Article ADS CAS Google Scholar
Colwell, R. Global climate and infectious disease: the cholera paradigm. Science 274 , 2025–2035 (1996).
Kim, J. & Ahn, I. Weekly ILI patient ratio change prediction using news articles with support vector machine. BMC Bioinform.. 20 , 1–16 (2019).
Article Google Scholar
Ghosh, S. et al. Temporal topic modeling to assess associations between news trends and infectious disease outbreaks. Sci. Rep. 7 , 40841 (2017).
Zhang, Y., Ibaraki, M. & Schwartz, F. W. Disease surveillance using online news: Dengue and zika in tropical countries. J. Biomed. Inform. 102 , 103374 (2020).
Charkraborty, S. & Subramanian, L. Extracting signals from news streams for disease outbreak prediction. In Proceedings of the IEEE Global Conference on Signal and Information Processing 1300–1304 (2016)
Steinberger, R., Fuart, F. & Best, C. et al. Text mining from the web for medical intelligence. Min. Massive Data Sets Secur. 19 , 295–310 (2008)
Shin, H. & Cho, S. Neighborhood property-based pattern selection for support vector machines. Neural Comput. 19 , 816–855 (2007).
Subramanya, A. & Bilmes, J. Soft-supervised learning for text classification. In Proceedings of the Conference on Empirical Methods in Natural Language Processing. Honolulu, Hawaii 1090–1099 (2008)
Allouche, O. et al. Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic. J. Appl. Ecol. 43 , 1223–1232 (2006).
Download references
Acknowledgements
This work was supported by a National Research Council of Science & Technology (NST) grant, funded by the Korea government (MSIP) (No. CRC-16-01-KRICT). This work was supported by the National Research Foundation of Korea (NRF) grant, funded by the Korea government (MEST) (No. 2016M3A9B6915714).
Author information
Authors and affiliations.
Department of Data-Centric Problem Solving Research, Korea Institute of Science and Technology Information, Yuseong-gu, Daejeon, Korea
Juhyeon Kim & Insung Ahn
Center for Convergent Research of Emerging Virus Infection, Korea Research Institute of Chemical Technology, Yuseong-gu, Daejeon, Korea
You can also search for this author in PubMed Google Scholar
Contributions
J.K. and I.A. conceptualized the study, and visualized the data and results. J.K. curated the data, performed formal analysis, validated the results, and authored the primary manuscript. I.A. administered and supervised the project, and also reviewed and edited the writing. All authors read and approved the final manuscript.
Corresponding author
Correspondence to Insung Ahn .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Kim, J., Ahn, I. Infectious disease outbreak prediction using media articles with machine learning models. Sci Rep 11 , 4413 (2021). https://doi.org/10.1038/s41598-021-83926-2
Download citation
Received : 30 March 2020
Accepted : 10 February 2021
Published : 24 February 2021
DOI : https://doi.org/10.1038/s41598-021-83926-2
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Machine learning approaches to identify systemic lupus erythematosus in anti-nuclear antibody-positive patients using genomic data and electronic health records.
- Chih-Wei Chung
- Seng-Cho Chou
- Yi-Ming Chen
BioData Mining (2024)
Deep learning techniques for detection and prediction of pandemic diseases: a systematic literature review
- Sunday Adeola Ajagbe
- Matthew O. Adigun
Multimedia Tools and Applications (2024)
Emerging infectious disease surveillance using a hierarchical diagnosis model and the Knox algorithm
- Mengying Wang
- Bingqing Yang
Scientific Reports (2023)
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: AI and Robotics newsletter — what matters in AI and robotics research, free to your inbox weekly.
Advertisement
Chronic Kidney Disease Prediction Using Machine Learning Techniques
- Original Paper
- Published: 31 August 2022
- Volume 1 , pages 534–540, ( 2023 )
Cite this article
- Saurabh Pal ORCID: orcid.org/0000-0001-9545-7481 1
6363 Accesses
11 Citations
Explore all metrics
Chronic kidney disease (CKD) is a life-threatening condition that can be difficult to diagnose early because there are no symptoms. The purpose of the proposed study is to develop and validate a predictive model for the prediction of chronic kidney disease. Machine learning algorithms are often used in medicine to predict and classify diseases. Medical records are often skewed. We have used chronic kidney disease dataset from UCI Machine learning repository with 25 features and applied three machine learning classifiers Logistic Regression (LR), Decision Tree (DT), and Support Vector Machine (SVM) for analysis and then used bagging ensemble method to improve the results of the developed model. The clusters of the chronic kidney disease dataset were used to train the machine learning classifiers. Finally, the Kidney Disease Collection is summarized by category and non-linear features. We get the best result in the case of decision tree with accuracy of 95.92%. Finally, after applying the bagging ensemble method we get the highest accuracy of 97.23%.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
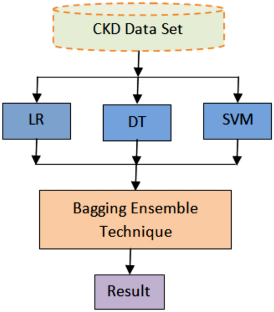
Similar content being viewed by others

Artificial intelligence in disease diagnosis: a systematic literature review, synthesizing framework and future research agenda
Yogesh Kumar, Apeksha Koul, … Muhammad Fazal Ijaz
Heart Disease Prediction using Machine Learning Techniques
Devansh Shah, Samir Patel & Santosh Kumar Bharti
Machine Learning in Healthcare Analytics: A State-of-the-Art Review
Surajit Das, Samaleswari P. Nayak, … Sarat Chandra Nayak
Aljaaf, A.J. 2018 Early Prediction of Chronic Kidney Disease Using Machine Learning Supported by Predictive Analytics. In Proceedings of the IEEE Congress on Evolutionary Computation (CEC). Wellington. New Zealand
A. Nishanth, T. Thiruvaran, Identifying important attributes for early detection of chronic kidney disease. IEEE Rev. Biomed. Eng. 11 , 208–216 (2018)
Article Google Scholar
A. Ogunleye, Q.-G. Wang, XGBoost model for chronic kidney disease diagnosis. IEEE/ACM Trans. Comput. Biol. Bioinform. 17 , 2131–2140 (2020)
F. Aqlan, R. Markle, A. Shamsan, "Data mining for chronic kidney disease prediction." in IIE Annual Conference. Proceedings, Institute of Industrial and Systems Engineers , (IISE 2017), pp. 1789–1794
N. Borisagar, D. Barad, P. Raval, Chronic kidney disease prediction using back propagation neural network algorithm. Proce. Int. Confe. Commun. Netw. 19–20 , 295–303 (2017)
Google Scholar
C. Bemando, E. Miranda, M. Aryuni, "Machine-Learning-Based Prediction Models of Coronary Heart Disease Using Naïve Bayes and Random Forest Algorithms," in 2021 International Conference on Software Engineering & Computer Systems and 4th International Conference on Computational Science and Information Management (ICSECS-ICOCSIM) , (IEEE, 2021), pp. 232–237
R.P. Ram Kumar, SanjeevaPolepaka, Performance comparison of random forest classifier and convolution neural network in predicting heart diseases, in Proceedings of the Third International Conference on Computational Intelligence and Informatics . ed. by K. SrujanRaju, A. Govardhan, B. PadmajaRani, R. Sridevi, M. Ramakrishna Murty (Springer, Singapore, 2020)
H. Singh, N. V. Navaneeth, G. N. Pillai, "Multisurface proximal SVM based decision trees for heart disease classification," in TENCON 2019-2019 IEEE Region 10 Conference (TENCON) , (IEEE 2019), pp. 13–18
S.D. Desai, S. Giraddi, P. Narayankar, N.R. Pudakalakatti, S. Sulegaon, Backpropagation neural network versus logistic regression in heart disease classification in advanced computing and communication technologies (Springer, Singapore, 2019)
D.D. Patil, R.P. Singh, V.M. Thakare, A.K. Gulve, Analysis of ecg arrhythmia for heart disease detection using svm and cuckoo search optimized neural network. Int. J. Eng. Technol. 7 (217), 27–33 (2018)
N. Liu, Z. Lin, J. Cao, Z. Koh, T. Zhang, G.-B. Huang, W. Ser, M.E.H. Ong, An intelligent scoring system and its application to cardiac arrest prediction. IEEE Trans. Inf Technol. Biomed. 16 (6), 1324–1331 (2012)
U. Rajendra Acharya, Oh. Shu Lih, Y. Hagiwara, J.H. Tan, M. Adam, A. Gertych, R.S. Tan, A deep convolutional neural network model to classify heartbeats. Comput. Biol. Med. 89 , 389–396 (2017)
R.S. Walse, G.D. Kurundkar, S.D. Khamitkar, A.A. Muley, P.U. Bhalchandra, S.N. Lokhande, Effective use of naïve bayes, decision tree, and random forest techniques for analysis of chronic kidney disease, in International Conference on Information and Communication Technology for Intelligent Systems . ed. by T. Senjyu, P.N. Mahalle, T. Perumal, A. Joshi (Springer, Singpore, 2020)
A. Nithya, A. Appathurai, N. Venkatadri, D.R. Ramji, C.A. Palagan, Kidney disease detection and segmentation using artificial neural network and multi-kernel k-means clustering for ultrasound images. Measurement (2020). https://doi.org/10.1016/j.measurement.2019.106952
Abdullah Al Imran, Md Nur Amin, and Fatema Tuj Johora. Classification of chronic kidney disease using logistic regression, feedforward neural network and wide & deep learning. In 2018 International Conference on Innovation in Engineering and Technology (ICIET), pages 1–6. IEEE, 2018.
B. Navaneeth, M. Suchetha, A dynamic pooling based convolutional neural network approach to detect chronic kidney disease. Biomed. Signal Proce. Control 62 , 102068 (2020)
A. Brunetti, G.D. Cascarano, I. De Feudis, M. Moschetta, L. Gesualdo, V. Bevilacqua, Detection and segmentation of kidneys from magnetic resonance images in patients with autosomal dominant polycystic kidney disease, in International Conference on Intelligent Computing . ed. by D.-S. Huang, K.-H. Jo, Z.-K. Huang (Springer International Publishing, Cham, 2019)
D. Ramos et al., Using decision tree to select forecasting algorithms in distinct electricity consumption context of an office building. Energy Rep. 8 , 417–422 (2022)
H.E. Song et al., Predictive modeling of groundwater nitrate pollution and evaluating its main impact factors using random forest. Chemosphere 290 , 133388 (2022)
H.U. Rongyao et al., Multi-task multi-modality SVM for early COVID-19 diagnosis using chest CT data. Inf. Proc. Manag. 59 (1), 102782 (2022)
X.U. Ankun et al., Artificial neural network (ANN) modeling for the prediction of odor emission rates from landfill working surface. Waste Manag. 138 , 158–171 (2022)
D.C. Yadav, S. Pal, An Ensemble Approach on the behalf of Classification and Prediction of Diabetes Mellitus Disease Emerging Trends in Data Driven Computing and Communications (Springer, Singapore, 2021)
D.C. Yadav, S. Pal, Performance based evaluation of algorithms on chronic kidney disease using hybrid ensemble model in machine learning. Biomed. Pharmacol. J. 14 (3), 1633–1646 (2021)
D.C. Yadav, S. Pal, Discovery of Thyroid Disease Using Different Ensemble Methods with Reduced Error Pruning Technique, in Computer-aided Design and Diagnosis Methods on the behalf of Biomedical Applications . ed. by G.R. Varun Bajaj, V.B. Sinha, G.R. Sinha (CRC Press, Boca Raton, 2021)
A. Zoda et al., Inferring genetic characteristics of Japanese Black cattle populations using genome-wide single nucleotide polymorphism markers. J. Animal Genet. 50 (1), 3–9 (2022)
G.M. Ifraz, M.H. Rashid, T. Tazin, S. Bourouis, M.M. Khan, Comparative analysis for prediction of kidney disease using intelligent machine learning methods. Comput. Math. Methods Med. (2021). https://doi.org/10.1155/2021/6141470
S. Krishnamurthy, K.S. Kapeleshh, E. Dovgan, M. Luštrek, B.G. Piletič, K. Srinivasan, Y.C. Li, A. Gradišek, S. Syed-Abdul, "Machine learning prediction models for chronic kidney disease using national health insurance claim data in Taiwan." medRxiv. (2020). https://doi.org/10.1101/2020.06.25.20139147
Z.U. Rehman, M.S. Zia, G.R. Bojja, M. Yaqub, F. Jinchao, K. Arshid, Texture based localization of a brain tumor from MR-images by using a machine learning approach. Med. Hypotheses 141 , 109705 (2020)
X. Han, X. Zheng, Y. Wang, X. Sun, Y. Xiao, Y. Tang, W. Qin, Random forest can accurately predict the development of end-stage renal disease in immunoglobulin a nephropathy patient. Annals Transl. Med. (2019). https://doi.org/10.21037/atm.2018.12.11
E.H.A. Rady, A.S. Anwar, Prediction of kidney disease stages using data mining algorithms. Inform. Med. Unlocked (2019). https://doi.org/10.1016/j.imu.2019.100178
Z. Dong, Q. Wang, Y. Ke, W. Zhang, Q. Hong, C. Liu, X. Chen, Prediction of 3-year risk of diabetic kidney disease using machine learning based on electronic medical records. J. Transl. Med. 20 (1), 1–10 (2022)
D.C. Yadav, S. Pal, Prediction of thyroid disease using decision tree ensemble method. Human-Intell. Syst. Integr. 2 (1), 89–95 (2020)
V. Chaurasia, S. Pal, Applications of machine learning techniques to predict diagnostic breast cancer. SN Compu. Sci. 1 (5), 1–11 (2020)
Chaurasia, V., & Pal, S. (2014). Performance analysis of data mining algorithms for diagnosis and prediction of heart and breast cancer disease. Review of research. 3(8).
Download references
Acknowledgements
Author thanks to Veer Bahadur Singh Purvanchal University, Jaunpur for providing the support for conducting this research work as a part of minor project “Analysis of Hidden Pattern and Discover Real Fact of Medical Diseases using Integrated Machine Learning Techniques.
Author information
Authors and affiliations.
Department of Computer Applications, VBS Purvanchal University, Jaunpur, India
Saurabh Pal
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Saurabh Pal .
Ethics declarations
Conflict of interest.
Author declares no conflict of interest.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Pal, S. Chronic Kidney Disease Prediction Using Machine Learning Techniques. Biomedical Materials & Devices 1 , 534–540 (2023). https://doi.org/10.1007/s44174-022-00027-y
Download citation
Received : 20 April 2022
Accepted : 16 August 2022
Published : 31 August 2022
Issue Date : March 2023
DOI : https://doi.org/10.1007/s44174-022-00027-y
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Chronic kidney disease
- Decision Tree
- Support Vector Machine
- Logistic Regression and Bagging Ensemble Method
- Find a journal
- Publish with us
- Track your research
- Open access
- Published: 18 April 2024
The predictive power of data: machine learning analysis for Covid-19 mortality based on personal, clinical, preclinical, and laboratory variables in a case–control study
- Maryam Seyedtabib ORCID: orcid.org/0000-0003-1599-9374 1 ,
- Roya Najafi-Vosough ORCID: orcid.org/0000-0003-2871-5748 2 &
- Naser Kamyari ORCID: orcid.org/0000-0001-6245-5447 3
BMC Infectious Diseases volume 24 , Article number: 411 ( 2024 ) Cite this article
45 Accesses
1 Altmetric
Metrics details
Background and purpose
The COVID-19 pandemic has presented unprecedented public health challenges worldwide. Understanding the factors contributing to COVID-19 mortality is critical for effective management and intervention strategies. This study aims to unlock the predictive power of data collected from personal, clinical, preclinical, and laboratory variables through machine learning (ML) analyses.
A retrospective study was conducted in 2022 in a large hospital in Abadan, Iran. Data were collected and categorized into demographic, clinical, comorbid, treatment, initial vital signs, symptoms, and laboratory test groups. The collected data were subjected to ML analysis to identify predictive factors associated with COVID-19 mortality. Five algorithms were used to analyze the data set and derive the latent predictive power of the variables by the shapely additive explanation values.
Results highlight key factors associated with COVID-19 mortality, including age, comorbidities (hypertension, diabetes), specific treatments (antibiotics, remdesivir, favipiravir, vitamin zinc), and clinical indicators (heart rate, respiratory rate, temperature). Notably, specific symptoms (productive cough, dyspnea, delirium) and laboratory values (D-dimer, ESR) also play a critical role in predicting outcomes. This study highlights the importance of feature selection and the impact of data quantity and quality on model performance.
This study highlights the potential of ML analysis to improve the accuracy of COVID-19 mortality prediction and emphasizes the need for a comprehensive approach that considers multiple feature categories. It highlights the critical role of data quality and quantity in improving model performance and contributes to our understanding of the multifaceted factors that influence COVID-19 outcomes.
Peer Review reports
Introduction
The World Health Organization (WHO) has declared COVID-19 a global pandemic in March 2020 [ 1 ]. The first cases of SARSCoV-2, a new severe acute respiratory syndrome coronavirus, were detected in Wuhan, China, and rapidly spread to become a global public health problem [ 2 ]. The clinical presentation and symptoms of COVID-19 may be similar to those of Middle East Respiratory Syndrome (MERS) and Severe Acute Respiratory Syndrome (SARS), however the rate of spread is higher [ 3 ]. By December 31, 2022, the pandemic had caused more than 729 million cases and nearly 6.7 million deaths (0.92%) were confirmed in 219 countries worldwide [ 4 ]. For many countries, figuring out what measures to take to prevent death or serious illness is a major challenge. Due to the complexity of transmission and the lack of proven treatments, COVID-19 is a major challenge worldwide [ 5 , 6 ]. In middle- and low-income countries, the situation is even more catastrophic due to high illiteracy rates, a very poor health care system, and lack of intensive care units [ 5 ]. In addition, understanding the factors contributing to COVID-19 mortality is critical for effective management and intervention strategies [ 6 ].
Numerous studies have shown several factors associated with COVID-19 outcomes, including socioeconomic, environmental, individual demographic, and health factors [ 7 , 8 , 9 ]. Risk factors for COVID -19 mortality vary by study and population studied [ 10 ]. Age [ 11 , 12 ], comorbidities such as hypertension, cardiovascular disease, diabetes, and COPD [ 13 , 14 , 15 ], sex [ 13 ], race/ethnicity [ 11 ], dementia, and neurologic disease [ 16 , 17 ], are some of the factors associated with COVID-19 mortality. Laboratory factors such as elevated levels of inflammatory markers, lymphopenia, elevated creatinine levels, and ALT are also associated with COVID-19 mortality [ 5 , 18 ]. Understanding these multiple risk factors is critical to accurately diagnose and treat COVID-19 patients.
Accurate diagnosis and treatment of the disease requires a comprehensive assessment that considers a variety of factors. These factors include personal factors such as medical history, lifestyle, and genetics; clinical factors such as observations on physical examinations and physician reports; preclinical factors such as early detection through screening or surveillance; laboratory factors such as results of diagnostic tests and medical imaging; and patient-reported signs and symptoms. However, the variety of characteristics associated with COVID-19 makes it difficult for physicians to accurately classify COVID-19 patients during the pandemic.
In today's digital transformation era, machine learning plays a vital role in various industries, including healthcare, where substantial data is generated daily [ 19 , 20 , 21 ]. Numerous studies have explored machine learning (ML) and explainable artificial intelligence (AI) in predicting COVID-19 prognosis and diagnosis [ 22 , 23 , 24 , 25 ]. Chadaga et al. have developed decision support systems and triage prediction systems using clinical markers and biomarkers [ 22 , 23 ]. Similarly, Khanna et al. have developed a ML and explainable AI system for COVID-19 triage prediction [ 24 ]. Zoabi has also made contributions in this field, developing ML models that predict COVID-19 test results with high accuracy based on a small number of features such as gender, age, contact with an infected person and initial clinical symptoms [ 25 ]. These studies emphasize the potential of ML and explainable AI to improve COVID-19 prediction and diagnosis. Nonetheless, the efficacy of ML algorithms heavily relies on the quality and quantity of data utilized for training. Recent research has indicated that deep learning algorithms' performance can be significantly enhanced compared to traditional ML methods by increasing the volume of data used [ 26 ]. However, it is crucial to acknowledge that the impact of data volume on model performance can vary based on data characteristics and experimental setup, highlighting the need for careful consideration and analysis when selecting data for model training. While the studies emphasize the importance of features in training ML algorithms for COVID-19 prediction and diagnosis, additional research is required on methods to enhance the interpretability of features.
Therefore, the primary aim of this study is to identify the key factors associated with mortality in COVID -19 patients admitted to hospitals in Abadan, Iran. For this purpose, seven categories of factors were selected, including demographic, clinical and conditions, comorbidities, treatments, initial vital signs, symptoms, and laboratory tests, and machine learning algorithms were employed. The predictive power of the data was assessed using 139 predictor variables across seven feature sets. Our next goal is to improve the interpretability of the extracted important features. To achieve this goal, we will utilize the innovative SHAP analysis, which illustrates the impact of features through a diagram.
Materials and methods
Study population and data collection.
Using data from the COVID-19 hospital-based registry database, a retrospective study was conducted from April 2020 to December 2022 at Ayatollah Talleghani Hospital (a COVID‑19 referral center) in Abadan City, Iran.
A total of 14,938 patients were initially screened for eligibility for the study. Of these, 9509 patients were excluded because their transcriptase polymerase chain reaction (RT-PCR) test results were negative or unspecified. The exclusion of patients due to incomplete or missing data is a common issue in medical research, particularly in the use of electronic medical records (EMRs) [ 27 ]. In addition, 1623 patients were excluded because their medical records contained more than 70% incomplete or missing data. In addition, patients younger than 18 years were not included in the study. The criterion for excluding 1623 patients due to "70% incomplete or missing data" means that the medical records of these patients did not contain at least 30% of the data required for a meaningful analysis. This threshold was set to ensure that the dataset used for the study contained a sufficient amount of complete and reliable information to draw accurate conclusions. Incomplete or missing data in a medical record may relate to key variables such as patient demographics, symptoms, lab results, treatment information, outcomes, or other data points important to the research. Insufficient data can affect the validity and reliability of study results and lead to potential bias or inaccuracies in the findings. It is important to exclude such incomplete records to maintain the quality and integrity of the research findings and to ensure that the conclusions drawn are based on robust and reliable data. After these exclusions, 3806 patients remained. Of these patients, 474 died due to COVID -19, while the remaining 3332 patients recovered and were included in the control group. To obtain a balanced sample, the control group was selected with a propensity score matching (PSM). The PSM refers to a statistical technique used to create a balanced comparison group by matching individuals in the control group (in this case, the survived group) with individuals in the case group (in this case, the deceased group) based on their propensity scores. In this study, the propensity scores for each person represented the probability of death (coded as a binary outcome; survived = 0, deceased = 1) calculated from a set of covariates (demographic factors) using the matchit function from the MatchIt library. Two individuals, one from the deceased group and one from the survived group, are considered matched if the difference between their propensity scores is small. Non-matching participants are discarded. The matching aims to reduce bias by making the distribution of observed characteristics similar between groups, which ultimately improves the comparability of groups in observational studies [ 28 ]. In total, the study included 1063 COVID-19 patients who belonged to either the deceased group (case = 474) or the survived group (control = 589) (Fig. 1 ).
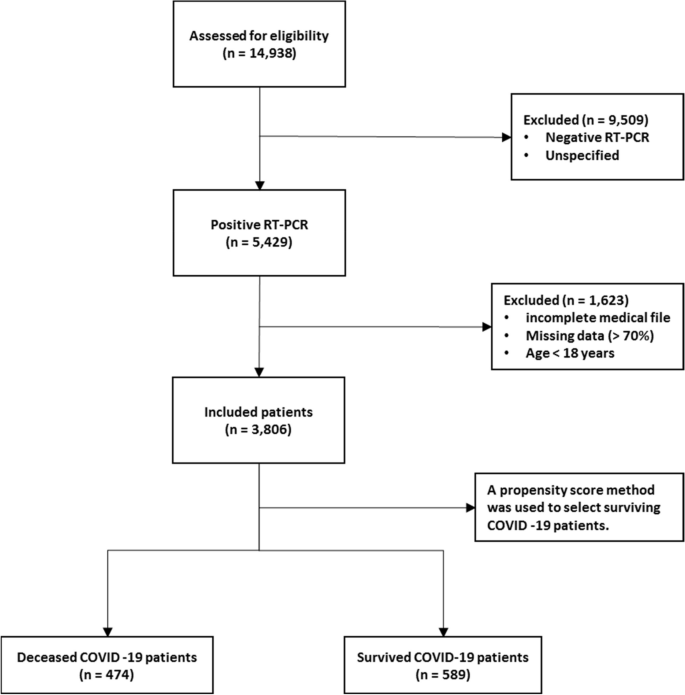
Flowchart describing the process of patient selection
In the COVID‑19 hospital‑based registry database, one hundred forty primary features in eight main classes including patient’s demographics (eight features), clinical and conditions features (16 features), comorbidities (18 features), treatment (17 features), initial vital sign (14 features), symptoms during hospitalization (31 features), laboratory results (35 features), and an output (0 for survived and 1 for deceased) was recorded for COVID-19 patients. The main features included in the hospital-based COVID-19 registry database are provided in Appendix Table 1 .
To ensure the accuracy of the recorded information, discharged patients or their relatives were called and asked to review some of the recorded information (demographic information, symptoms, and medical history). Clinical symptoms and vital signs were referenced to the first day of hospitalization (at admission). Laboratory test results were also referenced to the patient’s first blood sample at the time of hospitalization.
The study analyzed 140 variables in patients' records, normalizing continuous variables and creating a binary feature to categorize patients based on outcomes. To address the issue of an imbalanced dataset, the Synthetic Minority Over-sampling Technique (SMOTE) was utilized. Some classes were combined to simplify variables. For missing data, an imputation technique was applied, assuming a random distribution [ 29 ]. Little's MCAR test was performed with the naniar package to assess whether missing data in a dataset is missing completely at random (MCAR) [ 30 ]. The null hypothesis in this test is that the data are MCAR, and the test statistic is a chi-square value.
The Ethics Committee of Abadan University of Medical Science approved the research protocol (No. IR.ABADANUMS.REC.1401.095).
Predictor variables
All data were collected in eight categories, including demographic, clinical and conditions, comorbidities, treatment, initial vital signs, symptoms, and laboratory tests in medical records, for a total of 140 variables.
The "Demographics" category encompasses eight features, three of which are binary variables and five of which are categorical. The "Clinical Conditions" category includes 16 features, comprising one quantitative variable, 12 binary variables, and five categorical features. " Comorbidities ", " Treatment ", and " Symptoms " each have 18, 17, and 30 binary features, respectively. Also, there is one quantitative variable in symptoms category. The "Initial Vital Signs" category features 11 quantitative variables, two binary variables, and one categorical variable. Finally, the "Laboratory Tests" category comprises 35 features, with 33 being quantitative, one categorical, and one binary (Appendix Table 1 ).
Outcome variable
The primary outcome variable was mortality, with December 31, 2022, as the last date of follow‐up. The feature shows the class variable, which is binary. For any patient in the survivor group, the outcome is 0; otherwise, it is 1. In this study, 44.59% ( n = 474) of the samples were in the deceased group and were labeled 1.
Data balancing
In case–control studies, it is common to have unequal size groups since cases are typically fewer than controls [ 31 ]. However, in case–control studies with equal sizes, data balancing may not be necessary for ML algorithms [ 32 ]. When using ML algorithms, data balancing is generally important when there is an imbalance between classes, i.e., when one class has significantly fewer observations than the other [ 33 ]. In such cases, balancing can improve the performance of the algorithm by reducing the bias in favor of the majority class [ 34 ]. For case–control studies of the same size, the balance of the classes has already been reached and balancing may not be necessary. However, it is always recommended to evaluate the performance of the ML algorithm with the given data set to determine the need for data balancing. This is because unbalanced case–control ratios can cause inflated type I error rates and deflated type I error rates in balanced studies [ 35 ].
Feature selection
Feature selection is about selecting important variables from a large dataset to be used in a ML model to achieve better performance and efficiency. Another goal of feature selection is to reduce computational effort by eliminating irrelevant or redundant features [ 36 , 37 ]. Before generating predictions, it is important to perform feature selection to improve the accuracy of clinical decisions and reduce errors [ 37 ]. To identify the best predictors, researchers often compare the effectiveness of different feature selection methods. In this study, we used five common methods, including Decision Tree (DT), eXtreme Gradient Boosting (XGBoost), Support Vector Machine (SVM), Naïve Bayes (NB), and Random Forest (RF), to select relevant features for predicting mortality of COVID -19 patients. To avoid overfitting, we performed ten-fold cross-validation when training our dataset. This approach may help ensure that our model is optimized for accurate predictions of health status in COVID -19 patients.
Model development, evaluation, and clarity
In this study, the predictive models were developed with five ML algorithms, including DT, XGBoost, SVM, NB, and RF, using the R programming language (v4.3.1) and its packages [ 38 ]. We used cross-validation (CV) to tune the hyperparameters of our models based on the training subset of the dataset. For training and evaluating our ML models, we used a common technique called tenfold cross validation [ 39 ]. The primary training dataset was divided into ten folding, each containing 10% of the total data, using a technique called stratified random sampling. For each of the 30% of the data, a ML model was built and trained on the remaining 70% of the data. The performance of the model was then evaluated on the 30%-fold sample. This process was repeated 100 times with different training and test combinations, and the average performance was reported.
Performance measures include sensitivity (recall), specificity, accuracy, F1-score, and the area under the receiver operating characteristics curve (AUC ROC). Sensitivity is defined as TP / (TP + FN), whereas specificity is TN / (TN + FP). F1-score is defined as the harmonic mean of Precision and Recall with equal weight, where Precision equals TP + TN / total. Also, AUC refers to the area under the ROC curve. In the evaluation of ML techniques, values were classified as poor if below 50%, ok if between 50 and 80%, good if between 80 and 90%, and very good if greater than 90%. These criteria are commonly used in reporting model evaluations [ 40 , 41 ].
Finally, the shapely additive explanation (SHAP) method was used to provide clarity and understanding of the models. SHAP uses cooperative game theory to determine how each feature contributes to the prediction of ML models. This approach allows the computation of the contribution of each feature to model performance [ 42 , 43 ]. For this purpose, the package shapr was used, which includes a modified iteration of the kernel SHAP approach that takes into account the interdependence of the features when computing the Shapley values [ 44 ].

Patient characteristics
Table 1 shows the baseline characteristics of patients infected with COVID-19, including demographic data such as age and sex and other factors such as occupation, place of residence, marital status, education level, BMI, and season of admission. A total of 1063 adult patients (≥ 18 years) were enrolled in the study, of whom 589 (55.41%) survived and 474 (44.59%) died. Analysis showed that age was significantly different between the two groups, with a mean age of 54.70 ± 15.60 in the survivor group versus 65.53 ± 15.18 in the deceased group ( P < 0.001). There was also a significant association between age and survival, with a higher proportion of patients aged < 40 years in the survivor group (77.0%) than in the deceased group (23.0%) ( P < 0.001). No significant differences were found between the two groups in terms of sex, occupation, place of residence, marital status, and time of admission. However, there was a significant association between educational level and survival, with a lower proportion of patients with a college degree in the deceased group (37.2%) than in the survivor group (62.8%) ( P = 0.017). BMI also differed significantly between the two groups, with the proportion of patients with a BMI > 30 (kg/cm 2 ) being higher in the deceased group (56.5%) than in the survivor group (43.5%) ( P < 0.001).
Clinical and conditions
Important insights into the various clinical and condition characteristics associated with COVID-19 infection outcomes provides in Table 2 . The results show that patients who survived the infection had a significantly shorter hospitalization time (2.20 ± 1.63 days) compared to those who died (4.05 ± 3.10 days) ( P < 0.001). Patients who were admitted as elective cases had a higher survival rate (84.6%) compared to those who were admitted as urgent (61.3%) or emergency (47.4%) cases. There were no significant differences with regard to the number of infections or family infection history. However, patients who had a history of travel had a lower decease rate (40.1%).
A significantly higher proportion of deceased patients had cases requiring CPR (54.7% vs. 45.3%). Patients who had underlying medical conditions had a significantly lower survival rate (38.3%), with hyperlipidemia being the most prevalent condition (18.7%). Patients who had a history of alcohol consumption (12.5%), transplantation (30.0%), chemotropic (21.4%) or special drug use (0.0%), and immunosuppressive drug use (30.0%) also had a lower survival rate. Pregnant patients (44.4%) had similar survival outcomes compared to non-pregnant patients (55.6%). Patients who were recent or current smokers (36.4%) also had a significantly lower survival rate.
Comorbidities
Table 3 summarizes the comorbidity characteristics of COVID-19 infected patients. Out of 1063 patients, 54.84% had comorbidities. Chi-Square tests for individual comorbidities showed that most of them had a significant association with COVID-19 outcomes, with P -values less than 0.05. Among the various comorbidities, hypertension (HTN) and diabetes mellitus (DM) were the most prevalent, with 12% and 11.5% of patients having these conditions, respectively. The highest fatality rates were observed among patients with cardiovascular disease (95.5%), chronic kidney disease (62.5%), gastrointestinal (GI) (93.3%), and liver diseases (73.3%). Conversely, patients with neurology comorbidities had the lowest fatality rate (0%). These results highlight the significant role of comorbidities in COVID-19 outcomes and emphasize the need for special attention to be paid to patients with pre-existing health conditions.
The treatment characteristics of the COVID-19 patients and the resulting outcomes are shown in Table 4 . The table shows the frequency of patients who received different types of medications or therapies during their treatment. According to the results, the use of antibiotics (35.1%), remdesivir (29.6%), favipiravir (36.0%), and Vitamin zinc (33.5%) was significantly associated with a lower mortality rate ( P < 0.001), suggesting that these medications may have a positive impact on patient outcomes. On the other hand, the use of Heparin (66.1%), Insulin (82.6%), Antifungal (89.6%), ACE inhibitors (78.1%), and Angiotensin II Receptor Blockers (ARB) (83.8%) was significantly associated with increased mortality ( P < 0.001), suggesting that these medications may have a negative effect on the patient's outcome. Also, It seems that taking hydroxychloroquine (51.0%) is associated with a worse outcome at lower significance ( P = 0.022). The use of Atrovent, Corticosteroids and Non-Steroidal Anti-Inflammatory Drugs (NSAIDs) did not show a significant association with survival or mortality rates. Similarly, the use of Intravenous Immunoglobulin (IVIg), Vitamin C, Vitamin D, and Diuretic did not show a significant association with the patient’s outcome.
Initial vital signs
Table 5 provides initial vital sign characteristics of COVID-19 patients, including heart rate, respiratory rate, temperature, blood pressure, oxygen therapy, and radiography test result. The findings shows that deceased patients had higher HR (83.03 bpm vs. 76.14 bpm, P < 0.001), lower RR (11.40 bpm vs. 16.25 bpm, P < 0.001), higher temperature (37.43 °C vs. 36.91 °C, P < 0.001), higher SBP (128.16 mmHg vs. 123.33 mmHg, P < 0.001), and higher O 2 requirements (invasive: 75.0% vs. 25.0%, P < 0.001) compared to the survived patients. Additionally, deceased patients had higher MAP (99.35 mmHg vs. 96.08 mmHg, P = 0.005), and lower SPO 2 percentage (81.29% vs. 91.95%, P < 0.001) compared to the survived patients. Furthermore, deceased patients had higher PEEP levels (5.83 cmH2O vs. 0.69 cmH2O, P < 0.001), higher FiO2 levels (51.43% vs. 8.97%, P < 0.001), and more frequent bilateral pneumonia (63.0% vs. 37.0%, P < 0.001) compared to the survived patients. There appears to be no relationship between diastolic blood pressure and treatment outcome (83.44 mmHg vs. 85.61 mmHg).
Table 6 provides information on the symptoms of patients infected with COVID-19 by survival outcome. The table also shows the frequency of symptoms among patients. The most common symptom reported by patients was fever, which occurred in 67.0% of surviving and deceased patients. Dyspnea and nonproductive cough were the second and third most common symptoms, reported by 40.4% and 29.3% of the total sample, respectively. Other common symptoms listed in the Table were malodor (28.7%), dyspepsia (28.4%), and myalgia (25.6%).
The P -values reported in the table show that some symptoms are significantly associated with death, including productive cough, dyspnea, sore throat, headache, delirium, olfactory symptoms, dyspepsia, nausea, vomiting, sepsis, respiratory failure, heart failure, MODS, coagulopathy, secondary infection, stroke, acidosis, and admission to the intensive care unit. Surviving and deceased patients also differed significantly in the average number of days spent in the ICU. There was no significant association between patient outcomes and symptoms such as nonproductive cough, chills, diarrhea, chest pain, and hyperglycemia.
Laboratory tests
Table 7 shows the laboratory values of COVID-19 patients with the average values of the different laboratory results. The results show that the deceased patients had significantly lower levels of red blood cells (3.78 × 106/µL vs. 5.01 × 106/µL), hemoglobin (11.22 g/dL vs. 14.10 g/dL), and hematocrit (34.10% vs. 42.46%), whereas basophils and white blood cells did not differ significantly between the two groups. The percentage of neutrophils (65.59% vs. 62.58%) and monocytes (4.34% vs. 3.93%) was significantly higher in deceased patients, while the percentage of lymphocytes and eosinophils did not differ significantly between the two groups. In addition, deceased patients had higher levels of certain biomarkers, including D-dimer (1.347 mgFEU/L vs. 0.155 mgFEU/L), lactate dehydrogenase (174.61 U/L vs. 128.48 U/L), aspartate aminotransferase (93.09 U/L vs. 39.63 U/L), alanine aminotransferase (74.48 U/L vs. 28.70 U/L), alkaline phosphatase (119.51 IU/L vs. 81.34 IU/L), creatine phosphokinase-MB (4.65 IU/L vs. 3.33 IU/L), and positive troponin I (56.5% vs. 43.5%). The proportion of patients with positive C-reactive protein was also higher in the deceased group.
Other laboratory values with statistically significant differences between the two groups ( P < 0.001) were INR, ESR, BUN, Cr, Na, K, P, PLT, TSH, T3, and T4. The surviving patients generally had lower values in these laboratory characteristics than the deceased patients.
Model performance and evaluation
Five ML algorithms, namely DT, XGBoost, SVM, NB, and RF, were used in this study to build mortality prediction models COVID -19. The models were based on the optimal feature set selected in a previous step and were trained on the same data set. The effectiveness of the models was evaluated by calculating sensitivity, specificity, accuracy, F1 score, and AUC metrics. Table 8 shows the results of this performance evaluation. The average values are expressed from the test set as the mean (standard deviation).
The results show that the performance of the models varies widely in the different feature categories. The Laboratory Tests category achieved the highest performance, with all models scoring 100% in all metrics. The Symptoms and initial Vital Signs categories also show high performance, with XGBoost achieving the highest accuracy of 98.03% and DT achieving the highest sensitivity of 92.79%.
The Clinical and Conditions category also showed high performance, with all models showing accuracy above 91%. XGBoost achieved the highest sensitivity and specificity of 92.74% and 92.96%, respectively. In contrast, the Demographics category showed the lowest performance, with all models achieving less than 66.5% accuracy.
In summary, the results suggest that certain feature categories may be more useful than others in predicting mortality from COVID-19 and that some ML models may perform better than others depending on the feature category used.
Feature importance
SHapley Additive exPlanations (SHAP) values indicate the importance or contribution of each feature in predicting model output. These values help to understand the influence and importance of each feature on the model's decision-making process.
In Fig. 2 , the mean absolute SHAP values are shown to depict global feature importance. Figure 2 shows the contribution of each feature within its respective group as calculated by the XGBoost prediction model using SHAP. According to the SHAP method, the features that had the greatest impact on predicting COVID-19 mortality were, in descending order: D-dimer, CPR, PEEP, underlying disease, ESR, antifungal treatment, PaO2, age, dyspnea, and nausea.
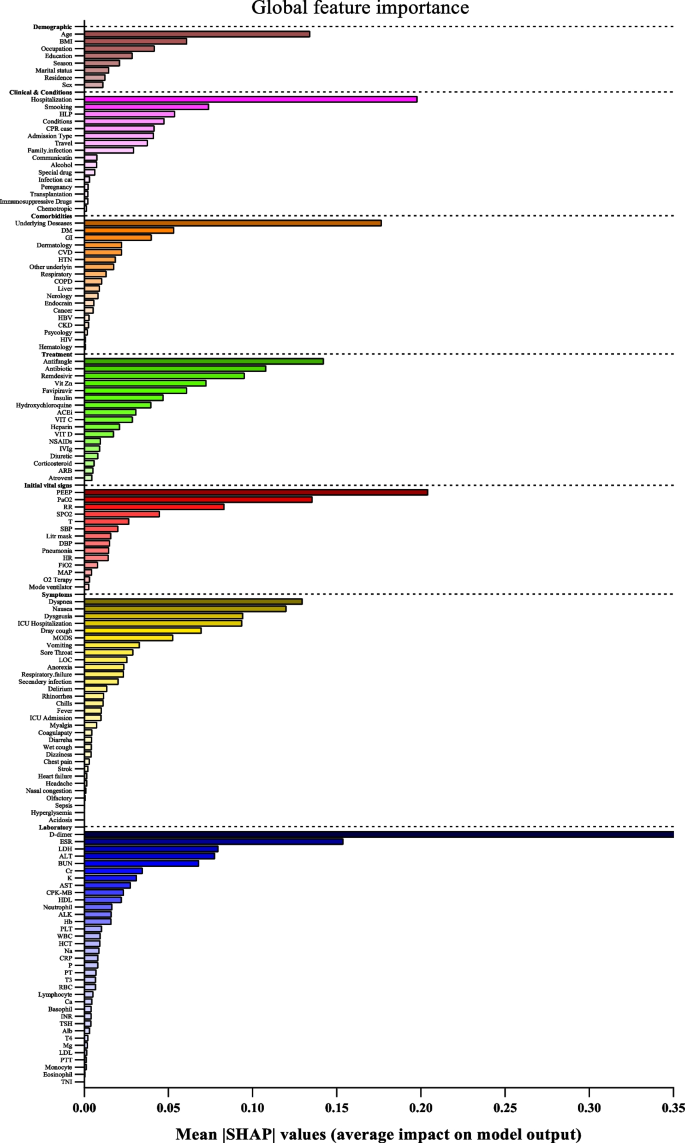
Feature importance based on SHAP-values. The mean absolute SHAP values are depicted, to illustrate global feature importance. The SHAP values change in the spectrum from dark (higher) to light (lower) color
On the other hand, Fig. 3 presents the local explanation summary that indicates the direction of the relationship between a variable and COVID-19 outcome. As shown in Fig. 3 (I to VII), older age and very low BMI were the two demographic factors with the greatest impact on model outcome, followed by clinical factors such as higher CPR, hospitalization, and hyperlipidemia. Higher mortality rates were associated with patients who smoked and had traveled in the past 14 days. Patients with underlying diseases, especially HTN, died more frequently. In contrast, the use of remdesivir, Vit Zn, and favipiravir is associated with lower mortality. Initial vital signs such as high PEEP, low PaO2 and RR had the greatest impact, as did symptoms such as dyspnea, MODS, sore throat and LOC. A higher risk of mortality is observed in patients with higher D-dimer levels and ESR as the most consequential laboratory tests, followed by K, AST and CPK-MB.
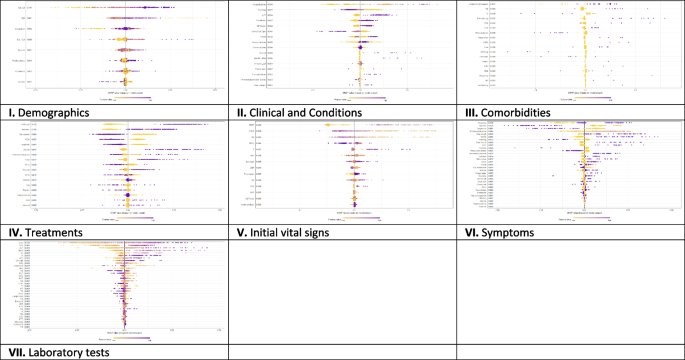
The SHAP-based feature importance of all categories (I to VII) for COVID‑19 mortality prediction, calculated with the XGBoost model. The local explanatory summary shows the direction of the relationship between a feature and patient outcome. Positive SHAP values indicate death, whereas negative SHAP values indicate survival. As the color scale shows, higher values are blue while lower values are orenge
Using the feature types listed in Appendix Table 1 , Fig. 4 shows that the performance of ML algorithms can be improved by increasing the number of features used in training, especially in distinguishing between symptoms, comorbidities, and treatments. In addition, the amount and quality of data used for training can significantly affect algorithm performance, with laboratory tests being more informative than initial vital signs. Regarding the influence of features, quantitative features tend to have a more positive effect on performance than qualitative features; clinical conditions tend to be more informative than demographic data. Thus, both the amount of data and the type of features used have a significant impact on the performance of ML algorithms.
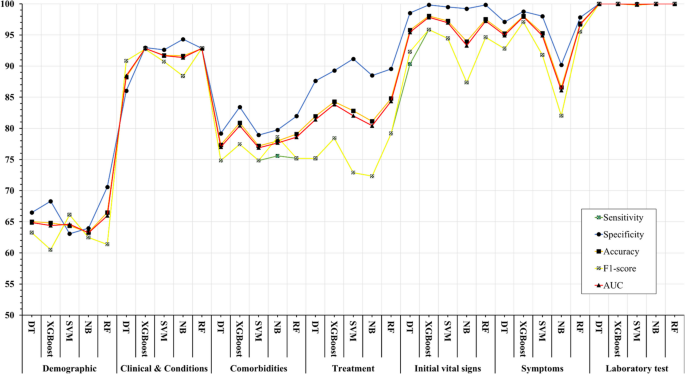
Association between feature sets and performance of machine learning algorithms in predicting COVID-19’s mortality
The COVID-19 pandemic has presented unprecedented public health challenges worldwide and requires a deep understanding of the factors contributing to COVID-19 mortality to enable effective management and intervention. This study used machine learning analysis to uncover the predictive power of an extensive dataset that includes wide range of personal, clinical, preclinical, and laboratory variables associated with COVID-19 mortality.
This study confirms previous research on COVID-19 outcomes that highlighted age as a significant predictor of mortality [ 45 , 46 , 47 ], along with comorbidities such as hypertension and diabetes [ 48 , 49 ]. Underlying conditions such as cardiovascular and renal disease also contribute to mortality risk [ 50 , 51 ].
Regarding treatment, antibiotics, remdesivir, favipiravir, and vitamin zinc are associated with lower mortality [ 52 , 53 ], whereas heparin, insulin, antifungals, ACE, and ARBs are associated with higher mortality [ 54 ]. This underscores the importance of drug choice in COVID -19 treatment.
Initial vital signs such as heart rate, respiratory rate, temperature, and oxygen therapy differ between surviving and deceased patients [ 55 ]. Deceased patients often have increased heart rate, lower respiratory rate, higher temperature, and increased oxygen requirements, which can serve as early indicators of disease severity.
Symptoms such as productive cough, dyspnea, and delirium are significantly associated with COVID-19 mortality, emphasizing the need for immediate monitoring and intervention [ 56 ]. Laboratory tests show altered hematologic and biochemical markers in deceased patients, underscoring the importance of routine laboratory monitoring in COVID-19 patients [ 57 , 58 ].
The ML algorithms were used in the study to predict mortality COVID-19 based on these multilayered variables. XGBoost and Random Forest performed better than other algorithms and had high recall, specificity, accuracy, F1 score, and AUC. This highlights the potential of ML, particularly the XGBoost algorithm, in improving prediction accuracy for COVID-19 mortality [ 59 ]. The study also highlighted the importance of drug choice in treatment and the potential of ML algorithms, particularly XGBoost, in improving prediction accuracy. However, the study's findings differ from those of Moulaei [ 60 ], Nopour [ 61 ], and Mehraeen [ 62 ] in terms of the best-performing ML algorithm and the most influential variables. While Moulaei [ 60 ] found that the random forest algorithm had the best performance, Nopour [ 61 ] and Ikemura [ 63 ] identified the artificial neural network and stacked ensemble models, respectively, as the most effective. Additionally, the most influential variables in predicting mortality varied across the studies, with Moulaei [ 60 ] highlighting dyspnea, ICU admission, and oxygen therapy, and Ikemura [ 63 ] identifying systolic and diastolic blood pressure, age, and other biomarkers. These differences may be attributed to variations in the datasets, feature selection, and model training.
However, it is important to note that the choice of algorithm should be tailored to the specific dataset and research question. In addition, the results suggest that a comprehensive approach that incorporates different feature categories may lead to more accurate prediction of COVID-19 mortality. In general, the results suggest that the performance of ML models is influenced by the number and type of features in each category. While some models consistently perform well across different categories (e.g., XGBoost), others perform better for specific types of features (e.g., SVM for Demographics).
Analysis of the importance of characteristics using SHAP values revealed critical factors affecting model results. D-dimer values, CPR, PEEP, underlying diseases, and ESR emerged as the most important features, highlighting the importance of these variables in predicting COVID-19 mortality. These results provide valuable insights into the underlying mechanisms and risk factors associated with severe COVID-19 outcomes.
The types of features used in ML models fall into two broad categories: quantitative (numerical) and qualitative (binary or categorical). The performance of ML methods can vary depending on the type of features used. Some algorithms work better with quantitative features, while others work better with qualitative features. For example, decision trees and random forests work well with both types of features [ 64 ], while neural networks often work better with quantitative features [ 65 , 66 ]. Accordingly, we consider these levels for the features under study to better assess the impact of the data.
The success of ML algorithms depends largely on the quality and quantity of the data on which they are trained [ 67 , 68 , 69 ]. Recent research, including the 2021 study by Sarker IH. [ 26 ], has shown that a larger amount of data can significantly improve the performance of deep learning algorithms compared to traditional machine learning techniques. However, it should be noted that the effect of data size on model performance depends on several factors, such as data characteristics and experimental design. This underscores the importance of carefully and judiciously selecting data for training.
Limitations
One of the limitations of this study is that it relies on data collected from a single hospital in Abadan, Iran. The data may not be representative of the diversity of COVID -19 cases in different regions, and there may be differences in data quality and completeness. In addition, retrospectively collected data may have biases and inaccuracies. Although the study included a substantial number of COVID -19 patients, the sample size may still limit the generalizability of the results, especially for less common subgroups or certain demographic characteristics.
Future works
Future studies could adopt a multi-center approach to improve the scope and depth of research on COVID-19 outcomes. This could include working with multiple hospitals in different regions of Iran to ensure a more diverse and representative sample. By conducting prospective studies, researchers can collect data in real time, which reduces the biases associated with retrospective data collection and increases the reliability of the results. Increasing sample size, conducting longitudinal studies to track patient progression, and implementing quality assurance measures are critical to improving generalizability, understanding long-term effects, and ensuring data accuracy in future research efforts. Collectively, these strategies aim to address the limitations of individual studies and make an important contribution to a more comprehensive understanding of COVID-19 outcomes in different populations and settings.
Conclusions
In summary, this study demonstrates the potential of ML algorithms in predicting COVID-19 mortality based on a comprehensive set of features. In addition, the interpretability of the models using SHAP-based feature importance, which revealed the variables strongly correlated with mortality. This study highlights the power of data-driven approaches in addressing critical public health challenges such as the COVID-19 pandemic. The results suggest that the performance of ML models is influenced by the number and type of features in each feature set. These findings may be a valuable resource for health professionals to identify high-risk patients COVID-19 and allocate resources effectively.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
World Health Organization
Middle east respiratory syndrome
Severe acute respiratory syndrome
Reverse transcription polymerase chain reaction
Propensity score matching
Synthetic minority over-sampling technique
Missing completely at random
Decision tree
EXtreme gradient boosting
Support vector machine
Naïve bayes
Random forest
Cross-validation
True positive
True negative
False positive
False negative
- Machine learning
Artificial Intelligence
Shapely additive explanation
Cardiopulmonary Resuscitation
Hypertension
Diabetes mellitus
Cardiovascular disease
Chronic Kidney disease
Chronic obstructive pulmonary disease
Human immunodeficiency virus
Hepatitis B virus
Such as influenza, pneumonia, asthma, bronchitis, and chronic obstructive airways disease
Gastrointestinal
Such as epilepsy, learning disabilities, neuromuscular disorders, autism, ADD, brain tumors, and cerebral palsy
Such as fatty liver disease and cirrhosis
Blood disease
Skin diseases
Mental disorders
Intravenous immunoglobulin
Non-steroidal anti-Inflammatory drugs
Angiotensin converting enzyme inhibitors
Angiotensin II receptor blockers
Beats per minute
Respiratory rate
Temperatures
Systolic blood pressure
Diastolic blood pressure
Mean arterial pressure
Oxygen saturation
Partial pressure of oxygen in the alveoli
Positive end-expiratory pressure
Fraction of inspired oxygen
Radiography (X-ray) test result
Smell disorders
Indigestion
Level of consciousness
Multiple organ dysfunction syndrome
Coughing up blood; Coagulopathy: bleeding disorder
High blood glucose
Intensive care unit
Red blood cell
White blood cell
Low-density lipoprotein
High-density lipoprotein
Prothrombin time
Partial thromboplastin time
International normalized ratio
Erythrocyte sedimentation rate
C-reactive-protein
Lactate dehydrogenase
Aspartate aminotransferase
Alanine aminotransferase
Alkaline phosphatase
Creatine phosphokinase-MB
Blood urea nitrogen
Thyroid stimulating hormone
Triiodothyronine
Coronavirus disease (COVID-19) pandemic. Available from: https://www.who.int/europe/emergencies/situations/covid-19 . [cited 2023 Sep 5].
Moolla I, Hiilamo H. Health system characteristics and COVID-19 performance in high-income countries. BMC Health Serv Res. 2023;23(1):1–14. https://doi.org/10.1186/s12913-023-09206-z . [cited 2023 Sep 5].
Article Google Scholar
Peeri NC, Shrestha N, Rahman MS, Zaki R, Tan Z, Bibi S, et al. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int J Epidemiol. 2020;49(3):717–26.
Article PubMed Google Scholar
WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard With Vaccination Data. Available from: https://covid19.who.int/ . [cited 2023 Sep 5].
Dessie ZG, Zewotir T. Mortality-related risk factors of COVID-19: a systematic review and meta-analysis of 42 studies and 423,117 patients. BMC Infect Dis. 2021;21(1):1–28. https://doi.org/10.1186/s12879-021-06536-3 . [cited 2023 Sep 5].
Article CAS Google Scholar
Wong ELY, Ho KF, Wong SYS, Cheung AWL, Yau PSY, Dong D, et al. Views on Workplace Policies and its Impact on Health-Related Quality of Life During Coronavirus Disease (COVID-19) Pandemic: Cross-Sectional Survey of Employees. Int J Heal Policy Manag. 2022;11(3):344–53. Available from: https://www.ijhpm.com/article_3879.html .
Google Scholar
Drefahl S, Wallace M, Mussino E, Aradhya S, Kolk M, Brandén M, et al. A population-based cohort study of socio-demographic risk factors for COVID-19 deaths in Sweden. Nat Commun. 2020;11(1):5097.
Article CAS PubMed PubMed Central Google Scholar
Islam N, Khunti K, Dambha-Miller H, Kawachi I, Marmot M. COVID-19 mortality: a complex interplay of sex, gender and ethnicity. Eur J Public Health. 2020;30(5):847–8.
Sarmadi M, Marufi N, Moghaddam VK. Association of COVID-19 global distribution and environmental and demographic factors: An updated three-month study. Environ Res. 2020;188:109748.
Aghazadeh-Attari J, Mohebbi I, Mansorian B, Ahmadzadeh J, Mirza-Aghazadeh-Attari M, Mobaraki K, et al. Epidemiological factors and worldwide pattern of Middle East respiratory syndrome coronavirus from 2013 to 2016. Int J Gen Med. 2018;11:121–5.
Risk of COVID-19-Related Mortality. Available from: https://www.cdc.gov/coronavirus/2019-ncov/science/data-review/risk.html . [cited 2023 Aug 26].
Bhaskaran K, Bacon S, Evans SJW, Bates CJ, Rentsch CT, MacKenna B, et al. Factors associated with deaths due to COVID-19 versus other causes: population-based cohort analysis of UK primary care data and linked national death registrations within the OpenSAFELY platform. Lancet Reg Heal. 2021;6:100-9.
Dessie ZG, Zewotir T. Mortality-related risk factors of COVID-19: a systematic review and meta-analysis of 42 studies and 423,117 patients. BMC Infect Dis. 2021;21(1):855. https://doi.org/10.1186/s12879-021-06536-3 .
Talebi SS, Hosseinzadeh A, Zare F, Daliri S, JamaliAtergeleh H, Khosravi A, et al. Risk Factors Associated with Mortality in COVID-19 Patient’s: Survival Analysis. Iran J Public Health. 2022;51(3):652–8.
PubMed PubMed Central Google Scholar
Singh J, Alam A, Samal J, Maeurer M, Ehtesham NZ, Chakaya J, et al. Role of multiple factors likely contributing to severity-mortality of COVID-19. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2021;96:105101.
CAS Google Scholar
Bhaskaran K, Bacon S, Evans SJ, Bates CJ, Rentsch CT, MacKenna B, et al. Factors associated with deaths due to COVID-19 versus other causes: population-based cohort analysis of UK primary care data and linked national death registrations within the OpenSAFELY platform. Lancet Reg Heal - Eur. 2021;6:100109. Available from: https://www.pmc/articles/PMC8106239/ . [cited 2023 Aug 26].
Ge E, Li Y, Wu S, Candido E, Wei X. Association of pre-existing comorbidities with mortality and disease severity among 167,500 individuals with COVID-19 in Canada: A population-based cohort study. PLoS One. 2021;16(10):e0258154. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0258154 . [cited 2023 Aug 26].
Tian S, Liu H, Liao M, Wu Y, Yang C, Cai Y, et al. Analysis of mortality in patients with COVID-19: clinical and laboratory parameters. Open Forum Infect Dis. 2020;7(5). Available from: https://dx.doi.org/10.1093/ofid/ofaa152 . [cited 2023 Aug 26].
Rashidi HH, Tran N, Albahra S, Dang LT. Machine learning in health care and laboratory medicine: General overview of supervised learning and Auto-ML. Int J Lab Hematol. 2021;43:15–22.
Najafi-Vosough R, Faradmal J, Hosseini SK, Moghimbeigi A, Mahjub H. Predicting hospital readmission in heart failure patients in Iran: a comparison of various machine learning methods. Healthc Inform Res. 2021;27(4):307–14.
Article PubMed PubMed Central Google Scholar
Alanazi A. Using machine learning for healthcare challenges and opportunities. Informatics Med Unlocked. 2022;100924:1–5.
Chadaga K, Prabhu S, Sampathila N, Chadaga R, Umakanth S, Bhat D, et al. Explainable artificial intelligence approaches for COVID-19 prognosis prediction using clinical markers. Sci Rep. 2024;14(1):1783.
Chadaga K, Prabhu S, Bhat V, Sampathila N, Umakanth S, Chadaga R, et al. An explainable multi-class decision support framework to predict COVID-19 prognosis utilizing biomarkers. Cogent Eng. 2023;10(2):2272361.
Khanna VV, Chadaga K, Sampathila N, Prabhu S, Chadaga R. A machine learning and explainable artificial intelligence triage-prediction system for COVID-19. Decis Anal J. 2023;100246:1–14.
Zoabi Y, Deri-Rozov S, Shomron N. Machine learning-based prediction of COVID-19 diagnosis based on symptoms. npj Digit Med. 2021;4(1):1–5.
IH Sarker 2021 Machine Learning: Algorithms, Real-World Applications and Research Directions SN Comput Sci. 2 3 160 Available from: https://doi.org/10.1007/s42979-021-00592-x .
Jones JA, Farnell B. Missing and Incomplete Data Reduces the Value of General Practice Electronic Medical Records as Data Sources in Research. Aust J Prim Health. 2007;13(1):74–80. Available from: https://www.publish.csiro.au/py/py07010 . [cited 2023 Dec 16].
Austin PC. An Introduction to Propensity Score Methods for Reducing the Effects of Confounding in Observational Studies. Multivariate Behav Res. 2011;46(3):399–424.
Torjusen H, Lieblein G, Næs T, Haugen M, Meltzer HM, Brantsæter AL. Food patterns and dietary quality associated with organic food consumption during pregnancy; Data from a large cohort of pregnant women in Norway. BMC Public Health. 2012;12(1):1–11.
Little RJA. A test of missing completely at random for multivariate data with missing values. J Am Stat Assoc. 1988;83(404):1198–202.
Tenny S, Kerndt CC, Hoffman MR. Case Control Studies. Encycl Pharm Pract Clin Pharm Vol 1-3 [Internet]. 2023;1–3:V2-356-V2-366. [cited 2024 Apr 14] Available from: https://www.ncbi.nlm.nih.gov/books/NBK448143/ .
Stanfill B, Reehl S, Bramer L, Nakayasu ES, Rich SS, Metz TO, et al. Extending Classification Algorithms to Case-Control Studies. Biomed Eng Comput Biol. 2019;10:117959721985895. Available from: https://www.pmc/articles/PMC6630079/ .[cited 2023 Sep 3].
Mulugeta G, Zewotir T, Tegegne AS, Juhar LH, Muleta MB. Classification of imbalanced data using machine learning algorithms to predict the risk of renal graft failures in Ethiopia. BMC Med Inform Decis Mak. 2023;23(1):1–17. https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-023-02185-5 . [cited 2023 Sep 3].
Sadeghi S, Khalili D, Ramezankhani A, Mansournia MA, Parsaeian M. Diabetes mellitus risk prediction in the presence of class imbalance using flexible machine learning methods. BMC Med Inform Decis Mak. 2022;22(1):36. https://doi.org/10.1186/s12911-022-01775-z .
Zhou W, Nielsen JB, Fritsche LG, Dey R, Gabrielsen ME, Wolford BN, et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet. 2018;50(9):1335. Available from: https://www.pmc/articles/PMC6119127/ . [cited 2023 Sep 3].
Miao J, Niu L. A Survey on Feature Selection. Procedia Comput Sci. 2016;91(1):919–26.
Remeseiro B, Bolon-Canedo V. A review of feature selection methods in medical applications. Comput Biol Med. 2019;112:103375.
Article CAS PubMed Google Scholar
R Studio Team. A language and environment for statistical computing. R Found Stat Comput. 2021;1.
Training Sets, Test Sets, and 10-fold Cross-validation - KDnuggets. Available from: https://www.kdnuggets.com/2018/01/training-test-sets-cross-validation.html . [cited 2023 Sep 4].
Hossin M, Sulaiman MN. A review on evaluation metrics for data classification evaluations. Int J data Min Knowl Manag Process. 2015;5(2):1.
Seyedtabib M, Kamyari N. Predicting polypharmacy in half a million adults in the Iranian population: comparison of machine learning algorithms. BMC Med Inform Decis Mak. 2023;23(1):84. https://doi.org/10.1186/s12911-023-02177-5 .
Lundberg SM, Lee S-I. A unified approach to interpreting model predictions. Adv Neural Inf Process Syst. 2017;30:4765–74.
Greenwell B. Fastshap: Fast approximate shapley values. Man R Packag v0 05. 2020;9–12. https://www.CRANR-projectorg/package=fastshap . Last accessed.
Aas K, Jullum M, Løland A. Explaining individual predictions when features are dependent: More accurate approximations to Shapley values. Artif Intell. 2021;298:103502.
Mesas AE, Cavero-Redondo I, Álvarez-Bueno C, Sarriá Cabrera MA, de Maffei Andrade S, Sequí-Dominguez I, et al. Predictors of in-hospital COVID-19 mortality: A comprehensive systematic review and meta-analysis exploring differences by age, sex and health conditions. PLoS One. 2020;15(11):e0241742.
Yanez ND, Weiss NS, Romand J-A, Treggiari MM. COVID-19 mortality risk for older men and women. BMC Public Health. 2020;20(1):1–7.
Sasson I. Age and COVID-19 mortality. Demogr Res. 2021;44:379–96.
Huang I, Lim MA, Pranata R. Diabetes mellitus is associated with increased mortality and severity of disease in COVID-19 pneumonia–a systematic review, meta-analysis, and meta-regression. Diabetes Metab Syndr Clin Res Rev. 2020;14(4):395–403.
Albitar O, Ballouze R, Ooi JP, Ghadzi SMS. Risk factors for mortality among COVID-19 patients. Diabetes Res Clin Pract. 2020;166:108293.
Di Castelnuovo A, Bonaccio M, Costanzo S, Gialluisi A, Antinori A, Berselli N, et al. Common cardiovascular risk factors and in-hospital mortality in 3,894 patients with COVID-19: survival analysis and machine learning-based findings from the multicentre Italian CORIST Study. Nutr Metab Cardiovasc Dis. 2020;30(11):1899–913.
Ssentongo P, Ssentongo AE, Heilbrunn ES, Ba DM, Chinchilli VM. Association of cardiovascular disease and 10 other pre-existing comorbidities with COVID-19 mortality: A systematic review and meta-analysis. PLoS ONE. 2020;15(8):e0238215.
Beran A, Mhanna M, Srour O, Ayesh H, Stewart JM, Hjouj M, et al. Clinical significance of micronutrient supplements in patients with coronavirus disease 2019: A comprehensive systematic review and meta-analysis. Clin Nutr ESPEN. 2022;48:167–77.
Perveen RA, Nasir M, Murshed M, Nazneen R, Ahmad SN. Remdesivir and favipiravir changes hepato-renal profile in COVID-19 patients: a cross sectional observation in Bangladesh. Int J Med Sci Clin Inven. 2021;8(1):5196–201.
El-Arif G, Khazaal S, Farhat A, Harb J, Annweiler C, Wu Y, et al. Angiotensin II Type I Receptor (AT1R): the gate towards COVID-19-associated diseases. Molecules. 2022;27(7):2048.
Ikram AS, Pillay S. Admission vital signs as predictors of COVID-19 mortality: a retrospective cross-sectional study. BMC Emerg Med. 2022;22(1):1–10.
Martí-Pastor A, Moreno-Perez O, Lobato-Martínez E, Valero-Sempere F, Amo-Lozano A, Martínez-García M-Á, et al. Association between Clinical Frailty Scale (CFS) and clinical presentation and outcomes in older inpatients with COVID-19. BMC Geriatr. 2023;23(1):1.
Lippi G, Plebani M. Laboratory abnormalities in patients with COVID-2019 infection. Clin Chem Lab Med. 2020;58(7):1131–4.
Naghashpour M, Ghiassian H, Mobarak S, Adelipour M, Piri M, Seyedtabib M, et al. Profiling serum levels of glutathione reductase and interleukin-10 in positive and negative-PCR COVID-19 outpatients: A comparative study from southwestern Iran. J Med Virol. 2022;94(4):1457–64.
Sharifi-Kia A, Nahvijou A, Sheikhtaheri A. Machine learning-based mortality prediction models for smoker COVID-19 patients. BMC Med Inform Decis Mak. 2023;23(1):1–15.
Moulaei K, Shanbehzadeh M, Mohammadi-Taghiabad Z, Kazemi-Arpanahi H. Comparing machine learning algorithms for predicting COVID-19 mortality. BMC Med Inform Decis Mak. 2022;22(1):2. https://doi.org/10.1186/s12911-021-01742-0 .
Nopour R, Erfannia L, Mehrabi N, Mashoufi M, Mahdavi A, Shanbehzadeh M. Comparison of Two Statistical Models for Predicting Mortality in COVID-19 Patients in Iran. Shiraz E-Medical J 2022 236 [Internet]. 2022;23(6):119172. [cited 2024 Apr 14] Available from: https://brieflands.com/articles/semj-119172 .
Mehraeen E, Karimi A, Barzegary A, Vahedi F, Afsahi AM, Dadras O, et al. Predictors of mortality in patients with COVID-19–a systematic review. Eur J Integr Med. 2020;40:101226.
Ikemura K, Bellin E, Yagi Y, Billett H, Saada M, Simone K, et al. Using Automated Machine Learning to Predict the Mortality of Patients With COVID-19: Prediction Model Development Study. J Med Internet Res [Internet]. 2021;23(2):e23458. Available from: https://www.jmir.org/2021/2/e23458 .
Breiman L. Random forests. Mach Learn. 2001;45:5–32.
Hinton G, Srivastava N, Swersky K. Neural networks for machine learning lecture 6a overview of mini-batch gradient descent. Cited on. 2012;14(8):2.
Zheng A, Casari A. Feature Engineering for Machine Learning: Principles and Techniques for Data Scientists. O’Reilly [Internet]. 2018;218. [cited 2024 Apr 14] Available from: https://www.amazon.com/Feature-Engineering-Machine-Learning-Principles/dp/1491953241 .
Adamson AS, Smith A. Machine Learning and Health Care Disparities in Dermatology. JAMA Dermatology. 2018;154(11):1247–8. Available from: https://jamanetwork.com/journals/jamadermatology/fullarticle/2688587 . [cited 2023 Sep 15].
Kavakiotis I, Tsave O, Salifoglou A, Maglaveras N, Vlahavas I, Chouvarda I. Machine Learning and Data Mining Methods in Diabetes Research. Comput Struct Biotechnol J. 2017;1(15):104–16.
Schmidt J, Marques MRG, Botti S, Marques MAL. Recent advances and applications of machine learning in solid-state materials science. Comput Mater. 2019;5(1):83. https://doi.org/10.1038/s41524-019-0221-0 .
Download references
Acknowledgements
We thank the Research Deputy of the Abadan University of Medical Sciences for financially supporting this project.
Summary points
∙ How can datasets improve mortality prediction using ML models for COVID-19 patients?
∙ In order, quantity and quality variables have more effect on the model performances.
∙ Intelligent techniques such as SHAP analysis can be used to improve the interpretability of features in ML algorithms.
∙ Well-structured data are critical to help health professionals identify at-risk patients and improve pandemic outcomes.
This research was supported by grant No. 1456 from the Abadan University of Medical Sciences. However, the funding source did not influence the study design, data collection, analysis and interpretation, report writing, or decision to publish the article.
Author information
Authors and affiliations.
Department of Biostatistics and Epidemiology, School of Health, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
Maryam Seyedtabib
Research Center for Health Sciences, Hamadan University of Medical Sciences, Hamadan, Iran
Roya Najafi-Vosough
Department of Biostatistics and Epidemiology, School of Health, Abadan University of Medical Sciences, Abadan, Iran
Naser Kamyari
You can also search for this author in PubMed Google Scholar
Contributions
MS: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Data curation, Writing–original draft, writing—review & editing, Visualization, Project administration. RNV: Conceptualization, Data curation, Formal analysis, Investigation, Writing–original draft, writing—review & editing. NK: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Resources, Data curation, Writing–original draft, writing—review & editing, Visualization, Supervision.
Corresponding author
Correspondence to Naser Kamyari .
Ethics declarations
Ethics approval and consent to participate.
This study was approved by the Research Ethics Committee (REC) of Abadan University of Medical Sciences under the ID number IR.ABADANUMS.REC.1401.095. Methods used complied with all relevant ethical guidelines and regulations. The Ethics Committee of Abadan University of Medical Sciences waived the requirement for written informed consent from study participants.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary material 1., rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Seyedtabib, M., Najafi-Vosough, R. & Kamyari, N. The predictive power of data: machine learning analysis for Covid-19 mortality based on personal, clinical, preclinical, and laboratory variables in a case–control study. BMC Infect Dis 24 , 411 (2024). https://doi.org/10.1186/s12879-024-09298-w
Download citation
Received : 22 December 2023
Accepted : 05 April 2024
Published : 18 April 2024
DOI : https://doi.org/10.1186/s12879-024-09298-w
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Predictive model
- Coronavirus disease
- Data quality
- Performance
BMC Infectious Diseases
ISSN: 1471-2334
- Submission enquiries: [email protected]
- General enquiries: [email protected]
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- J Healthc Eng
- v.2022; 2022

Identification and Prediction of Chronic Diseases Using Machine Learning Approach
Rayan alanazi.
Department of Computer Science, College of Science and Arts in Qurayyat, Jouf University, Sakakah, Saudi Arabia
Associated Data
The data used to support the findings of this study are included within the article.
Nowadays, humans face various diseases due to the current environmental condition and their living habits. The identification and prediction of such diseases at their earlier stages are much important, so as to prevent the extremity of it. It is difficult for doctors to manually identify the diseases accurately most of the time. The goal of this paper is to identify and predict the patients with more common chronic illnesses. This could be achieved by using a cutting-edge machine learning technique to ensure that this categorization reliably identifies persons with chronic diseases. The prediction of diseases is also a challenging task. Hence, data mining plays a critical role in disease prediction. The proposed system offers a broad disease prognosis based on patient's symptoms by using the machine learning algorithms such as convolutional neural network (CNN) for automatic feature extraction and disease prediction and K-nearest neighbor (KNN) for distance calculation to find the exact match in the data set and the final disease prediction outcome. A collection of disease symptoms has been performed for the preparation of the data set along with the person's living habits, and details related to doctor consultations are taken into account in this general disease prediction. Finally, a comparative study of the proposed system with various algorithms such as Naïve Bayes, decision tree, and logistic regression has been demonstrated in this paper.
1. Introduction
All over the world, chronic diseases are a critical issue in the healthcare domain. According to the medical statement, due to chronic diseases, the death rate of humans increases. The treatments given for this disease consume over 70% of the patient's income. Hence, it is highly essential to minimize the patient's risk factor that leads to death. The advancement in medical research makes health-related data collection easier [ 1 , 2 ]. The healthcare data includes the demographics, medical analysis reports, and the history of disease of the patient. The diseases caused could be varied based on the regions and the living habitats in that region. Hence, along with the disease data, the environmental condition and the living habitat of the patient should also be recorded in the data set.
In recent years, the healthcare domain is evolving more due to the integration of information technology (IT) in it. The intention to integrate IT in healthcare is to make the life of an individual more affordable with comfort as smartphones made one's life easier [ 3 ]. This could be possible by making healthcare to be intelligent, for instance, the invention of the smart ambulance, smart hospital facilities, and so on, which helps the patients and doctors in several ways [ 4 ]. The research on a specified region for patients affected by chronic diseases every year had been held and found that the difference between the patients in genderwise is very small, and it is found that the large number of patients were admitted in the year 2014 for treating chronic diseases. The use of structured and unstructured data provides highly accurate results instead of using only structured data. Since the unstructured data includes the doctor's records on the patients related to diseases and the patient's symptoms and grievances faced by them, explained by themselves, which is an added advantage when used along with the structured data that consists of the patient demographics, disease details, living habitats, and laboratory test results [ 5 , 6 ]. It is difficult to diagnose rare diseases. Hence, the use of self-reported behavioral data helps differentiate the individuals with rare diseases from the ones with common chronic diseases. By using machine learning approaches along with questionnaires, it is believed that the identification of rare diseases is highly possible [ 7 ].
In the last decade, some innovative technologies had been introduced to rapidly collect the data such as MRI (magnetic resonant imaging) readouts, ultrasonography, social media gained data, and electronically gained activity, behavioral, and clinical data. These big data sets of healthcare are high-dimensional, which means the number of features recorder per observation might be greater than the total observations. They are noisy, sparse, cross-sectional, and lacks statistical power. By applying machine learning techniques, the issues in the high-dimensional data sets can be overcome [ 8 ]. Machine learning contributes more in several domains. Many of the complex models make use of exiting larger training data, simultaneously at the edge of a major shift in healthcare epidemiology [ 9 ]. These data can enhance the knowledge gain in the risk factors of diseases to reduce healthcare-associated infections, improve patient risk stratification, and find the way of transmitting the infectious diseases [ 10 ]. Machine learning can facilitate the analysis of laboratory results and other details of patients for the early detection of diseases. The low-level data could be converted to high-level knowledge via knowledge discovery in the database so as to gain knowledge about the disease patterns to support early detection [ 11 ]. The data collected for creating a data set should be preprocessed for its missing values, and then only the important features needed for accurate disease prediction are selected so as to enhance the prediction accuracy and minimizing the time taken for model training [ 12 ].
In the era of the Internet and technologies, people are not concerned about their health and lives. As everyone is interested in surfing and social media activities, they ignore visiting hospitals for their health checkup. By taking this activity as an advantage, a machine learning model that takes the symptoms given as input and predicts the possibility and risk of the disease affected or the development of such diseases in an individual should be developed [ 13 , 14 ]. The more common chronic diseases are diabetes, cardiovascular diseases, cancer, strokes, hepatitis C, and arthritis. As these diseases persist for a long time and have a high mortality rate, the diagnosis of such diseases is highly important in the healthcare domain. Foreseeing the disease can help take preventive actions and avoid getting affected by it, and early detection of it can help provide better treatment [ 15 ]. There are various techniques in machine learning such as supervised, semisupervised, unsupervised, reinforcement, evolutionary, and deep learning. The problem is associated with the processing of extracted features from real data and structured as vectors [ 16 ]. The processing quality is based on the proper combination of those vectors. But, most of the times, the high dimensionality of the vectors or the discrepancies in the data makes a big issue. Hence, it is important to reduce the dimensionality of the data set even if it leads to a little loss of details to make the data set a highly compatible dimension. This reduction in the dimensionality of the data set improves the model performance [ 17 ].
The system of chronic diseases management is essential for those affected by such diseases and in need of proper medical assessment and treatment information [ 18 ]. Also, this system can be useful for individuals who are in need of self-care to improve their health condition, since it is proved that self-management is the primary care of those with chronic diseases, and it is considered as the unavoidable part of treatment. With the use of mobile applications, the health information of patients can be recorded, and they serve as a better tool to enable self-management [ 19 ]. To effectively predict a disease, information such as narration about the symptoms felt by the patients, details of consultation with medical practitioners, lab examination results, and computed tomography and X-ray images [ 20 ]. Little research is performed in identifying the accuracy and predictive power for developing a machine learning model with only information from lab examination results for the diagnosis of diseases. And, for performance enhancement, ensemble machine learning and deep learning model can be used [ 21 , 22 ]. In the healthcare domain, artificial intelligence (AI) plays a major role in automating the roles involved in disease diagnosis and treatment suggestions and also schedules perfect timing by the medical practitioners to perform various obligations that cannot be automated [ 23 ].
The major objective of the proposed system is to identify and predict chronic disease in an individual using a machine learning approach [ 24 , 25 ]. The data set comprises both the structured data that includes the patient's age, gender, height, weight, and so on, excluding the patient's personal information such as name and ID, and the unstructured data that includes the patient's symptoms, information related to consultation about the disease with the doctors, and the living habits of that individual [ 26 ]. These data are preprocessed for finding the missing values. They are then reconstructed to increase the quality of the model, thereby increasing the prediction accuracy. For prediction, the machine learning algorithms such as CNN and KNN are used [ 27 , 28 ]. This paper is organized as the details of the related works carried out while doing the research are given in Section 2 , the preliminaries of the algorithms used in given in Section 3 , the description of the proposed methodology in Section 4 , the result and discussion part are given in Section 5 followed by the conclusion in Section 6 , and finally, a list of references used in this study has been given.
2. Related Work
This section describes the related works that are performed in developing the proposed model for predicting chronic diseases. The following are the discussions made by reviewing the existing literature that helps develop the proposed system efficiently and effectively.
The objective variable of the study in [ 29 ] is the resource consumption such as medical and long-term care expenses and a predictive model for medical care using a random forest machine learning algorithm [ 30 ]. This method uses data of more than 100 pieces that includes preventive activities, clinical tests, and medical practices. This model uses mean decrease Gini for classification and for regression mean square error (MSE) is used [ 31 , 32 ]. The training model uses a grid search for hyperparameter tuning and is validated using K -fold cross-validation. Along with the objective variable, exploratory variables such as age, gender, and analysis period are also included, since the aim of this paper is proper management of the budget for medical care [ 33 ]. A review that highlights the applications of machine learning techniques in various medical practices such as predicting, diagnosing, and prognosis of diseases such as multiple sclerosis, autoimmune chronic kidney disease, autoimmune rheumatic disease, and inflammatory bowel disease and for the selection of treatments and stratification of patients; drug development; drug repurposing; target interpretation; and validation has been given in [ 34 , 35 ]. This paper also provides a detailed description of the challenges faced by the machine learning approaches such as the need for quality data in preparation of robust models, external model validation using the independent data set, difficulties faced during implementation of a model, and ethical concerns. A predictive model for chronic kidney disease is explained in [ 36 , 37 ]. This model is developed using four machine learning approaches such as support vector machine (SVM), logistic regression (LR), decision tree (DT), and KNN for classification purposes. The data set used in this paper is the Indian chronic kidney disease (CKD) that consists of 400 occurrences, 24 features, and 2 classes obtained from the UCI machine learning repository. The developed model is evaluated using a 5-fold cross-validation process, and the experiment is conducted on the Weka data mining tool and MATLAB and finally concluded that the SVM classifier attains higher accuracy when compared to the others.
A system that can predict multiple diseases with the help of various machine learning algorithms such as Naïve Bayes, KNN, DT, random forest, and SVM algorithms has been described in [ 38 ] to bridge the gap among the patients and the doctors to achieve their own goals. The existing approaches in the field of automatic disease prediction lack the patient's trust in the model's prediction and also reduce the need for doctors, which makes the doctors get panic about their livelihood. But this method integrates a module for doctor recommendation that solves both the issues by making sure the patient to trust due to the intervention of doctors and also improves the business of doctors. A model called PARAMO, which is a platform of a parallel predictive model that uses electronic health records (EHR) for healthcare analysis, has been implemented in [ 39 ]. This method comprises three phases, namely, the generation of the dependency graph, which removes redundancy and identifies dependency; then execution engine for dependency graph, which includes prioritizing, scheduling, and parallel execution; and finally the parallelization infrastructure. The PARAMO model is tested with three sets of real data, that is, small, medium, and large data sets that includes the medications, diagnosis data, and lab records, obtained from EHR that ranges from 5,000 to around 300,000 patients. In addition to this, the small and large sets include the procedure data, and the medium set includes the symptoms of heart failure that are taken from medical records [ 40 ]. An efficient recommendation system for chronic disease diagnosis has been demonstrated in [ 41 ]. This method uses a data mining approach. The data set used in this system includes medical data and two-dimensional data. The medical data include the data obtained from sensors or medical data entries, and the two-dimensional data include the external user and the item features. For enhancing the accuracy of prediction, the decision tree approach, which is a highly prevalent data mining approach, is used for classification. Various decision tree classifiers such as random forest, REP tree, decision stump, and J48 are involved in the creation of this predictive model. This model is tested with randomly selected 20 samples and found that the RF outperforms the other three algorithms.
Prediction of 3 types of immune diseases such as allergy, infectious, and autoimmune diseases using decision tree, maximum margin learning, and instance-based learning, respectively, has been given in [ 42 ]. The correlation between the classification of immunogens and its physicochemical properties is one of the purposes of this study. The immunogen data such as the stats of diseases, responses from B-cell, discontinuous epitope location, host, source organisms, and so on are collected from Immune Epitope Database (IEDB) and analyzed its 6 physicochemical properties such as PSSM (position-specific scoring matrix) information per position, hydrophilic scale, flexibility, antigenic propensity, hydropathy index, and side chain polarity. This system is tested using a method called leave-one-out cross-validation for the performance of prediction outcomes with parameters such as accuracy and F-score. A risk prediction model for predicting disease risks using a random forest machine learning approach from highly imbalance data has been described in [ 43 ]. The data set used in this approach is the Nationwide Inpatient Sample (NIS), which includes 8 million records of hospital stays with 126 clinical as well as nonclinical data. The nonclinical data comprises patient's demographics, hospital location, date and year of admission, pin code, treatment/diagnosis cost, and duration of stay in a hospital ward. The clinical data comprises the treatment procedures, its categories, diagnosis categories, and its codes. Each record has a vector containing 15 diagnosis codes characterized by International Classification of Diseases, 9 th Revision, Clinical Modification (ICD-9-CM). As the unbalance data produces undesirable results, a repeated random sampling method is employed to solve this issue. The developed model is evaluated using SVM, RF ensemble learning, bagging, and boosting algorithms. The study [ 44 ] demonstrates a novel adaptive probabilistic divergence-based feature selection algorithm to predict chronic kidney disease in its earlier stage. This algorithm is based on statistical and divergence information theory. For classification, the hyperparameterized logistic regression model is used in this study. The data set used in this approach is obtained from various hospitals and laboratories with information of 630 patients with 52 attributes, and this data set is given to the physician for verification of its correctness. The model developed is evaluated using the data sets of diabetes, heart, and kidney diseases, and the performance evaluation metrics followed in this study is the precision, recall, F1-score, and ROC (receiver operating characteristics) curve.
A system that enhances the risk prediction of a patient's health condition using a deep learning approach on big data and a revised fusion node model has been demonstrated in [ 45 ]. This deep learning model for extracting the data and logical inference is made of the combination of complex machine learning algorithm such as Bayesian fusion and neural networks. The architecture of this system consists of five layers, namely, the data layer that is responsible for data collection, data aggregation layer for data acquisition from several data sources and desired format changing, analytics layer to do proper analytics on the data aggregated, information exploration layer to create the output that makes the results of analytics understandable for users, and big data governance layer that is responsible for managing the above layers. Also, in this paper, the application of MapReduce is discussed for optimizing the analytics efficiency and also inspires the design of SOA (service-oriented architecture) for making the external systems easily access the results from analytics. A machine learning model of disease prediction cost has been implemented in [ 46 ] that uses big data, which includes structured and unstructured data for preparing the data set and the developed model is made available at affordable. The prediction algorithm used in this method is the decision tree algorithm and the MapReduce algorithm is applied for enhancing the efficiency of the operation. The advantages of this model are reduction in retrieval time of queries, improved accuracy. A method of predicting the risk of chronic kidney disease using zub machine learning approaches has been described in [ 47 , 48 ]. Two types of data sets are used in this method. One is from UCI with 400 instances and 35 features, and the other is a real-time data set obtained from Khulna City Medical College with 55 instances and 25 features. Data processing is done using Pandas and Numpy libraries, and the missing data are handled using median filtering. Feature extraction is performed using the Chi-square test. Model evaluation is performed using 10-fold cross-validation. Artificial neural network (ANN) and random forest algorithms are used for disease classification. This method is believed that it can predict the risk of chronic kidney disease in its earlier stage [ 49 – 52 ].
3. Preliminaries
3.1. chronic disease.
According to US National Center for Health Statistics, chronic diseases are diseases that last for a long period of time, that is, more than three months. These diseases are neither treated by medicines nor prevented by vaccines. The major cause of chronic diseases is the use of tobacco, unhealthy food habits, and lack of physical activity. Also, this disease can commonly be caused due to ageing. Chronic diseases include cardiovascular disease, cancer, arthritis, diabetes, obesity, epilepsy and seizures, and problems in oral health [ 35 ].
Cardiovascular disease includes heart disease and stroke, which highly lead to death. This disease is caused due to the use of tobacco, intake of nutritionless food, and lack of physical activity. When these activities are changed by the patient, they might have the chance to reduce the impact on controlling and preventing cardiovascular disease.
Next to cardiovascular disease, cancer such as colon cancer and breast cancer is considered the deadliest disease. It can be controlled only by prevention, early detection, and proper medical support. Minimizing the prevalence of environmental and behavioral factors that causes cancer reduces the chance risk of causing it.
The chronic disease such as arthritis causes inflammation in the joints, causes pain, and stiffness that increases due to ageing. There is an availability of cost-effective methods for reducing the effects caused by arthritis but are not used much. The effects of arthritis can be reduced by following moderate exercises regularly.
Diabetes is a serious and high-money-consuming disease. The impact of diabetes can be reduced by self-care and early detection of the disease [ 53 ]. Around 7 million people over the age of 65 or above are affected by this disease particularly type 2 diabetes.
Since 1980, obesity is more common in adults for all age groups. The one who is overweight or obese can develop the risk of getting high blood pressure (BP), heart diseases, diabetes, and arthritis. Obesity can also cause some types of cancers.
Epilepsy and seizures are highly costly in treatment [ 54 ]. This disease is common among all age groups, especially in young and elders.
Oral health problems are a crucial issue that attains special attention in the health of older people. This is a serious issue, since it affects the normal day-to-day actions of a person such as speak, chew, swallow, and maintain a nutritional food plan.
3.2. Convolutional Neural Network (CNN)
The ConvNet or CNN is an algorithm of deep learning that fetches the input and assigns the bias and weights to its several aspects and then distinguishes one from the other [ 55 ] as shown in Algorithm 1 . The major reason for using CNN is that it requires only few efforts in preprocessing the data when compared with other algorithms, since the CNN can learn to optimize the filters through automate learning [ 56 ]. The output layer of CNN can be calculated using the following expression:

Convolutional neural network algorithm.
3.3. K-Nearest Neighbor (KNN)
KNN is a supervised machine learning algorithm, which analyzes the similarities between the new data and the existing data and adds the new data into the category that is highly similar to the available categories [ 57 ] as shown in Algorithm 2 . The KNN can be used in classification as well as regression tasks, but it is most commonly used in classification. This algorithm is also called the lazy learner algorithm; since it will not learn instantly from the training data, it stores the data set and does its action during the classification process. The calculation of Euclidean distance is expressed mathematically as follows:

K-nearest neighbor algorithm.
4. Proposed Methodology
In this section, a detailed description of the data set creation, model preparation, and disease prediction has been given. The first action is data collection. Our proposed system collects structured and unstructured data obtained from various sources. After data collection, they are subjected to preprocessing and are split into cleaning and test data sets. Then the training data set is trained with the machine learning algorithms such as CNN and KNN to a number of epochs for improving the accuracy of the prediction results. After multiple epochs, once the desired target is achieved, the developed model is ready for testing.
At this step, the model is tested with the test data set to verify the model performance with brand-new data that were not used for training. If the model attains the desired accuracy in test data, then the proposed model is ready for deployment as shown in Figure 1 .

Architecture of proposed disease and risk prediction system.
4.1. Data Collection
The real-life data that includes structured data such as patient basic information including demographics, living habitat, and lab test results and the unstructured data such as the symptoms of the disease faced by the patient and their consultation with the doctor. The data set excludes the patient's personal details such as name, ID, and location so as to preserve their privacy.
4.2. Preprocecssing
The collected data are preprocessed for the availability of missing values in most of the structured data. Hence, it is essential to fill out the missed data or remove or modify them to enhance the quality of the data set. The preprocessing step also eliminates the commas, punctuations, and white spaces. Once the preprocessing of data has been completed, it is then subjected to feature extraction followed by disease prediction.
4.3. Model Description
As discussed above, the data set consists of both structured and unstructured data. The structured data comprises patient demographics and the data related to the cause for the disease such as age, gender height, weight, and so on, patient's living habitat, laboratory test results, and the disease that they are affected in tabular format. The unstructured data comprises patient's disease symptoms and the information about the interrogation with doctors in text format. The unstructured data is an added advantage of the prediction task to get a more accurate results. The data set is split into 80% for training and 20% for testing.
4.4. Disease Prediction Using CNN
The proposed system uses the CNN algorithm in the prediction of chronic disease. At first, the data set is converted into vector form, followed by word embedding to adopt zero values for filling the data. It is then given to the convolution layer.
The pooling layer takes the input from the convolution layer and follows the max pooling operation. The output of max pooling is given to the fully connected layer, and then finally, the output layer provides the classification results. Figure 2 shows the block diagram of the convolutional neural network.

Block diagram of convolutional neural network.
4.5. Distance Calculation Using KNN
In K -Nearest Neighbor (KNN), the value of K is known, and the features that are similar to the K value are called the nearest neighbor. The nearest neighbor to the known K value is chosen, and the nearest distance between them is calculated. The feature with less distance value is considered to be the exact match, which is the final disease prediction output. In the proposed system, Euclidean distance is used, since the result obtained by it is better when compared to other distance calculation methods. It is a nonparametric algorithm since it will not take decisions on original data. In KNN, the training input data are located in X and Y axes, and the test data are located in the plots of X and Y axes. Then, the plots of test data with less distance are chosen and are considered as the desired target. It is important to choose the value of the nearest K point should be always odd.
The calculation of Euclidean distance can be performed by using the following formula and is represented in Figure 3 :

Calculation of Euclidean distance.
5. Performance Evaluation
For evaluating the proposed disease prediction model, four performance evaluation metrics are used. The confusion matrix consists of the true positives (TP), which is the correct prediction of the target as a patient with chronic disease; the true negatives (TN), which is the correct prediction of the persons without diseases; false positives (FP), which is the incorrect prediction of the healthy person as a diseased person, and false negatives (FN), which is the incorrect prediction of the target as healthy persons. The following is the description of the four performance evaluation parameters.
5.1. Accuracy
The classification accuracy is described as the ratio of correct predicted values to the total predicted values and is depicted mathematically as follows:
5.2. Precision
The precision or positive predictive value (PPV) is described as the ratio of correct prediction to the total correct values including the true and false predictions and is depicted mathematically as follows:
5.3. Recall
The recall or sensitivity or true positive rate (TPR) is described as the ratio of correct predicted values to the sum of correct positive predictions and the incorrect negative predicted values and is depicted mathematically as follows:
5.4. F1-Score
The F-measure ( F β ) is described as the weighted average of the values obtained from the calculation of precision and recall parameters. Whenever the distribution of class is not even, then the value of F 1 − Score is highly important than the accuracy value. And whenever the values of false positives and negatives are dissimilar, the value of F 1 − Score is highly suitable. The F 1 − Score is depicted mathematically as follows:
By simplifying using β =1,
The obtained values of precision, recall, and F1-score of the proposed CNN and KNN model is compared with the values of the performance metrics of Naïve Bayes, decision tree, and logistic regression algorithms, and the results are tabulated in Table 1.
The accuracy is the important parameter since the prediction result is the important factor for the patient, and if it is wrong, then it will be a detriment to them. The other parameters such as precision, recall, and F1-score are for the evaluation of the model performance as shown in Table 1 .
Performance evaluation comparison.
Figure 4 shows the graphical representation of the comparison results of accuracies of the proposed and other algorithms. This graph illustrates the variations in the prediction accuracies of the four algorithms such as the Naïve Bayes, decision tree, logistic regression, and the proposed CNN and KNN algorithms as 52%, 62%, 86%, and 96%, respectively. This shows that the proposed system achieves the highest accuracy of 96% when compared to the other machine learning algorithms.

Comparison of accuracies of proposed and other algorithms.
Figure 5 shows the graphical representation of the comparison precision, recall, and F1-score values of the proposed and other algorithms. This graph illustrates the variations in the three performance evaluation parameters of the four algorithms such as the Naïve Bayes, decision tree, logistic regression, and the proposed CNN and KNN algorithms as 52%, 64%, 84%, and 93%, respectively, for precision; 80%, 605, 88%, and 99%, respectively, for recall; and 65%, 62%, 82%, and 97%, respectively, for F1-score. These results shows that the prosed model developed using CNN and KNN algorithm is considered to be the best of the remaining three algorithms with 93%, 99%, and 97% for precision, recall, and F1-score, respectively, which is higher when compared to the others.

Comparison of other performance evaluation metrics of proposed and other algorithms.
6. Conclusion
This paper proposed a method of identification and prediction of the presence of chronic disease in an individual using the machine learning algorithms such as CNN and KNN. The advantage of the proposed system is the use of both structured and unstructured data from real life for data set preparation, which lacks in many of the existing approaches. In this paper, the performance of the proposed model is compared with other algorithms such as Naïve Bayes, decision tree, and logistic regression algorithms. The results show that the proposed system provides an accuracy of 95% that is higher than that of the other two algorithms. It is highly believed that the proposed system can reduce the risk of chronic diseases by diagnosing them earlier and also reduces the cost for diagnosis, treatment, and doctor consultation.
Acknowledgments
This work was funded by the Deanship of Scientific Research at Jouf University under grant no. DSR-2021-02-0371.
Data Availability
Conflicts of interest.
The author declares that there are no conflicts of interest.
This paper is in the following e-collection/theme issue:
Published on 19.4.2024 in Vol 8 (2024)
Machine Learning–Based Prediction of Changes in the Clinical Condition of Patients With Complex Chronic Diseases: 2-Phase Pilot Prospective Single-Center Observational Study
Authors of this article:

Original Paper
- Celia Alvarez-Romero 1 , MSc ;
- Alejandro Polo-Molina 2 , MSc ;
- Eugenio Francisco Sánchez-Úbeda 2 , PhD ;
- Carlos Jimenez-De-Juan 3 , MSc ;
- Maria Pastora Cuadri-Benitez 3 , MSc ;
- Jose Antonio Rivas-Gonzalez 1 , BSc ;
- Jose Portela 2 , PhD ;
- Rafael Palacios 2 , PhD ;
- Carlos Rodriguez-Morcillo 2 , PhD ;
- Antonio Muñoz 2 , PhD ;
- Carlos Luis Parra-Calderon 1 , MSc ;
- Maria Dolores Nieto-Martin 3 , PhD ;
- Manuel Ollero-Baturone 3 , PhD ;
- Carlos Hernández-Quiles 3 , PhD
1 Computational Health Informatics Group, Institute of Biomedicine of Seville, Virgen del Rocío University Hospital, Consejo Superior de Investigaciones Científicas, University of, Seville, Spain
2 Institute for Research in Technology (IIT), ICAI School of Engineering, Comillas Pontifical University, Madrid, Spain
3 Internal Medicine Department, Virgen del Rocio University Hospital, Sevilla, Spain
Corresponding Author:
Carlos Hernández-Quiles, PhD
Internal Medicine Department
Virgen del Rocio University Hospital
Av Manuel Siurot s/n
Sevilla, 41013
Phone: 34 697950012
Email: [email protected]
Background: Functional impairment is one of the most decisive prognostic factors in patients with complex chronic diseases. A more significant functional impairment indicates that the disease is progressing, which requires implementing diagnostic and therapeutic actions that stop the exacerbation of the disease.
Objective: This study aimed to predict alterations in the clinical condition of patients with complex chronic diseases by predicting the Barthel Index (BI), to assess their clinical and functional status using an artificial intelligence model and data collected through an internet of things mobility device.
Methods: A 2-phase pilot prospective single-center observational study was designed. During both phases, patients were recruited, and a wearable activity tracker was allocated to gather physical activity data. Patients were categorized into class A (BI≤20; total dependence), class B (20<BI≤60; severe dependence), and class C (BI>60; moderate or mild dependence, or independent). Data preprocessing and machine learning techniques were used to analyze mobility data. A decision tree was used to achieve a robust and interpretable model. To assess the quality of the predictions, several metrics including the mean absolute error, median absolute error, and root mean squared error were considered. Statistical analysis was performed using SPSS and Python for the machine learning modeling.
Results: Overall, 90 patients with complex chronic diseases were included: 50 during phase 1 (class A: n=10; class B: n=20; and class C: n=20) and 40 during phase 2 (class B: n=20 and class C: n=20). Most patients (n=85, 94%) had a caregiver. The mean value of the BI was 58.31 (SD 24.5). Concerning mobility aids, 60% (n=52) of patients required no aids, whereas the others required walkers (n=18, 20%), wheelchairs (n=15, 17%), canes (n=4, 7%), and crutches (n=1, 1%). Regarding clinical complexity, 85% (n=76) met patient with polypathology criteria with a mean of 2.7 (SD 1.25) categories, 69% (n=61) met the frailty criteria, and 21% (n=19) met the patients with complex chronic diseases criteria. The most characteristic symptoms were dyspnea (n=73, 82%), chronic pain (n=63, 70%), asthenia (n=62, 68%), and anxiety (n=41, 46%). Polypharmacy was presented in 87% (n=78) of patients. The most important variables for predicting the BI were identified as the maximum step count during evening and morning periods and the absence of a mobility device. The model exhibited consistency in the median prediction error with a median absolute error close to 5 in the training, validation, and production-like test sets. The model accuracy for identifying the BI class was 91%, 88%, and 90% in the training, validation, and test sets, respectively.
Conclusions: Using commercially available mobility recording devices makes it possible to identify different mobility patterns and relate them to functional capacity in patients with polypathology according to the BI without using clinical parameters.
Introduction
The Spanish strategy for the approach to chronicity in the National Health System defines patients with complex chronic diseases as patients with 1 or more chronic diseases that present greater complexity in their management due to changing needs that force continuous evaluations and make necessary the coordinated use of various care levels and, in some cases, health and social [ 1 ]. Social changes and health advances mean that we are living longer and better and that most diseases affecting us are becoming chronic. Several of them are accumulating, which causes the growing phenomenon of people living with polypathology or complex chronic diseases. This concept includes not only people with the primary disease that triggers other secondary conditions but also those people where 2 or more chronic diseases coexist. It is a population characterized by frailty, polymedication, old age, hyperfrequent use of emergency services, and frequent re-entering. It is estimated that 70% to 95% of the older people in our environment have 1.2 to 4.2 chronic diseases, which constitute the leading death cause in the world (60% of the total) [ 2 ]. These patients generate a greater demand for attention in different care settings and use a more significant number of health and social resources. It is predominantly seen in older patients presenting with limiting and progressive diseases (eg, renal or cardiac insufficiency), polypharmacy, and some degree of functional impairment [ 3 ].
Functional impairment is one of the most decisive prognostic factors in patients with complex chronic diseases. A more significant functional impairment indicates that the disease is progressing, which requires implementing diagnostic and therapeutic actions that stop the exacerbation of the disease. The functional assessment of patients with complex chronic diseases can be performed using tools such as the Barthel Index (BI) [ 4 ], mobility tests, the 4-meter gait test [ 5 ], the balance test [ 6 ], and the timed “up and go” test [ 7 ].
The BI has excellent predictive value for variables such as mortality, hospital admission, and stay length in rehabilitation departments. In addition, it is an indicator to assess the functional and prognostic capacities of patients with complex chronic diseases [ 8 , 9 ]. The BI is a simple measure developed on empirical bases in obtaining and interpreting it. It is about assigning, to each patient, a score based on their degree of dependence to perform a series of basic activities related mainly to the individual’s mobility (eg, moving between the chair and the bed, moving, going up and down stairs, or showering). The total score can vary between 0 (fully dependent) and 100 points (completely independent) [ 10 ].
Concerning functional capacity, physical inactivity is defined as the spectrum of any decrease in body movement that reduces energy expenditure toward the baseline level. Physical inactivity affects many aspects of a person, such as respiratory capacity, bones, or the central nervous system, among others, and can even lead to various diseases [ 11 ]. In addition, physical inactivity itself decreases the physical fitness of the person, the duration of good health, and the age of onset of his or her first chronic illness. Relative to this, there are several parameters to assess the physical inactivity of the person, such as the number of daily steps, the time spent sitting, or the immobilization of the limbs, among others.
On the other hand, the possible causal relationship between sedentary behavior and mortality due to various causes has been studied. Various studies used accelerometers on the thigh to control the body’s position, and the chances of experiencing illnesses increased for every additional hour of sitting. Regarding limb immobilization in older people, one of the main concerns is the inability to recover the loss of bone strength and muscle mass [ 12 ].
Recent technological advances allow mobility monitoring through smartphones or wrist devices, which are widely distributed throughout the population. These devices provide information on the paths, the number of steps, the speed of the march, and the periods of falls, among others. Specific initiatives have tried to apply this information to the health sector. For instance, a multiagent system equipped with sensors has been developed to collect vital signs from patients. This system is intended to facilitate various tasks within the residences of older or disabled people [ 13 ]. Additionally, mobility monitoring by sensors in different rooms of the house has been considered to study translations between rooms and measure the length of stay in each room for older patients living alone [ 14 ].
Furthermore, individual physical activity can be monitored using accelerometers placed on the patient’s trunk and thigh [ 15 ]. At the same time, smartwatches have been used to evaluate movement and gait patterns in patients with Parkinson disease and essential tremor [ 16 ]. These advancements are driving the development of more hardware devices to enhance health care delivery and turn the concept of “a doctor in your pocket” into a reality for patients.
We would like to emphasize that using sensors to obtain health information currently has a specific trajectory [ 17 - 19 ]. Mobility has long presented prominent importance when dealing with diseases whose onset and symptomatic progression affect the functional capacity of the subject [ 20 ]. Recently, machine learning (ML) techniques are increasingly being considered to characterize the movement, or some particularities of the movement, which can provide relevant information about the patient’s clinical status [ 21 , 22 ]. Some works investigate the relationships between movement and specific clinical pathologies [ 23 , 24 ].
Despite these initiatives, the evaluation of the mobility of patients with complex chronic diseases and their relationship with the functional capacity measured by the BI has yet to be explored [ 25 ]. For all these reasons, and with this background, this study aims to develop and validate mobility patterns based on artificial intelligence and the internet of things (IoT) environment, aiming to predict changes in the clinical condition of patients with complex chronic diseases through the prediction of the BI to know the clinical and functional status of the patients.
Study Design and Recruitment
This 2-phase pilot observational study has been designed to analyze how mobility deterioration can reflect changes in the patient’s clinical condition and possible degeneration in the integrated care of patients with complex chronic diseases. To this end, a prospective, single-center, descriptive study was carried out.
Eligible patients met the criteria of chronic patients with complex health needs defined according to the Integrated Patient Care Process of the Andalusian Ministry of Health [ 26 ]. Concretely, the study population included patients older than 65 years of age with multimorbidity (ie, diagnosed with at least 2 chronic diseases), and the recruitment took place at the Virgen del Rocio University Hospital of Seville, Spain. In addition, those patients in a situation of agony or those whose vital prognosis was limited, patients with psychiatric disease, and patients or caregivers unable to use mobility devices were excluded from the study. The study subjects were patients of the Internal Medicine Department of the Virgen del Rocio University Hospital of Seville, as part of the Andalusian Health Service, Spain.
The research was conducted in 2 phases. In the initial phase (January to November 2022), a cohort of 50 patients was enrolled, and their BI was measured before the allocation of the wearable activity trackers (WATs), during routine doctor appointments after providing informed consent. Approximately 1 month after the first assessment (encounter 1), the BI was measured again (encounter 2) to evaluate any changes in their functional status ( Figure 1 ). The recruitment was conducted according to different degrees of patients with complex chronic diseases dependence based on the BI measured during the first assessment. In particular, the enrolled patient’s group was classified into 3 groups based on their BI scores: class A included patients with BI≤20 (total dependence), class B comprised patients with 20<BI≤60 (severe dependence), and class C consisted of patients with BI>60 (moderate or mild dependence, or independent) [ 27 ].

In the second phase (July 2022 to May 2023), 40 patients were recruited. Similar to phase 1, patients were recruited during doctor appointments, and their BI was measured 3 months after encounter 1 and encounter 2. Therefore, for phase 2 patients, there was an additional encounter 3.
An IoT framework was deployed to gather patient mobility data after analyzing the existing devices and applications in the market. The IoT-based infrastructure consisted of using mobile devices and WAT to measure the mobility activities of patients, considering the no or minimal invasion in the development of the daily tasks for the patients under study. The WAT used in this study recorded the step count, the cardiac activity, and the sleep duration from which both the step count and the heart rate were analyzed ( Figure 2 ).

Once the 90 patients were included in the study, they were assigned a WAT, and different mobility and functional status tests were conducted. An information system for data storage (Analytics Datastore) was developed. This database allowed both the dumping of the information collected through the WAT and the storage of the relevant clinical information of the patients extracted from the electronic health records ( Figure 3 ). For this purpose, confidentiality protocols of information and the security of the center’s systems were followed and in compliance with the ethical approval obtained by the hospital’s ethics committee.

The study patients’ exposure and clinical variables of interest were analyzed to characterize patient groups regarding mobility, using mobility measurement devices and clinical conditions. Demographic and clinical variables such as diseases, fragility, and polypathology criteria; pharmacological variables; and functional tests such as the BI, balance test, and timed “up and go” test were collected. Statistical analysis was performed using SPSS Statistics software (version 25; IBM Corp) and Python (version 3.10.9; Python Software Foundation) for the ML modeling.
Data Preparation for the ML Model
The WAT automatically gathered continuous and noninvasive data on a range of parameters, encompassing heart rates, step counts, and sleep duration. However, these raw data must be processed to apply ML techniques. Furthermore, given the potential influence of the walking aids on mobility patterns, patients were classified into 3 distinct groups. The first group encompassed patients using wheelchairs; the second comprised individuals using canes, walkers, or receiving aid from a caregiver during ambulation; and the third consisted of those with no reliance on assistance.
To ensure high data quality, instances where the median heart rate is missing are identified as null, along with the corresponding count of steps. The steps taken within 1-hour intervals are aggregated, and the median heart rate for these hourly intervals is computed. The resulting time series data of hourly step counts are then smoothed by applying a centered rolling window with a window size of 3. Subsequently, the data are grouped by the specific hour, resulting in an average representation of each patient’s activity throughout a 24-hour period. The data used to generate the mean activity profile consist of the information recorded during the 30-day period before encounter 2 requiring at least 14 days’ worth of data to consider the patient in the data set. This approach aims to develop a methodology that allows the estimation of the BI at any given moment using the information collected by the WAT over the last 30 days. Therefore, it holds the potential to provide a more dynamic and real-time assessment of the BI based on continuous monitoring through WAT. Additionally, mobility profiles from encounter 3 served as a production-like test set and were excluded from the model’s training. This strategy evaluated the model’s real-world performance and generalization on unseen data.
The 24-hour mean activity profiles were partitioned into 4-time segments: morning (7 AM-1 PM), afternoon (2 PM-7 PM), evening (8 PM-11 PM), and overnight (midnight-6 AM). This methodology aimed to reduce the dimensionality of the data inputted into the model. Various approaches were considered to reduce dimensionality, including summing the steps within each interval, calculating the mean, and determining the maximum value. The 4-time segments were selected based on the findings of Polo-Molina et al [ 25 ], where it was demonstrated that mobility patterns can be categorized into distinct clusters. The study highlighted that the maximum value of steps within each interval aligns with the suggested division, regardless of individual variations in the mobility patterns.
Once the data set was generated, it was divided into training and validation sets, with 70% (n=63) of the records allocated for training and 30% (n=27) for validation. To ensure that the proportions of each group of walking aids were maintained at this ratio, the division was performed within each group, and then the data were combined to create the final training and validation data sets. This approach aimed to ensure representative and well-balanced distributions of walking aids in both sets, allowing for robust evaluation of the model’s performance across different modes of mobility.
Explainable ML Model
A decision tree regressor has been considered to predict the BI. Decision trees iteratively select variables to maximize information gain or minimize impurity at each decision node, creating a hierarchical structure. Therefore, starting from the whole set of variables, at each split, the training algorithm selects the variable that generates the best split [ 28 ].
Moreover, to optimize the performance of the regression model, the hyperparameters were fine-tuned using a cross-validation approach with 7 folds. This technique ensures robustness and selects the optimal settings that yield the best predictive accuracy for the BI. The cross-validation optimization considered the hyperparameters “min_impurity_decrease” (ranging from 0.0 to 1.0 in increments of 0.01), “min_samples_leaf” (from 1 to 10 in steps of 1), and “min_samples_split” (spanning 1 to 10 with an interval of 1).
In addition, to assess the quality of the predictions, several metrics were considered, including the mean absolute error, median absolute error (MAD), and root mean squared error.
Finally, the permutation importance from explanatory variables was computed by permuting individual feature values while measuring the subsequent decline in model performance [ 28 ]. This iterative process assigned a score to each feature based on the decrease in predictive power caused by permutations, with elevated scores indicating significant contributions to accurate predictions.
Ethical Considerations
First, ethical approval was obtained in the health organization based on the regional regulations before involving it in the study execution. Likewise, informed consent procedures were defined, including informed consent and information sheets for the patients who were included in the study. Before starting this study, and based on the ethical and legal regulations, ethical approval was requested from the Ethics Committee of the Virgen del Rocio University Hospital of Seville, Spain. The study protocol, informed consent documents, and information sheets were submitted, and approval from the ethics committee was received. The study began, and patients who met the inclusion criteria were invited to participate after explaining the study procedures. Those who accepted and signed the informed consent and information sheets were included in the clinical study.
In addition, to ensure the protection of the privacy and confidentiality of the study participants, sensible data were anonymized and deidentified. Likewise, confidentiality protocols of information and the security of the center’s systems were followed and in compliance with the ethical approval obtained by the hospital’s ethics committee.
Statistical Analysis
A total of 90 patients were included in the study and were classified into 3 categories according to their BI. Concretely, 50 patients were enrolled in the first phase (10 in the BI class A, 20 in BI class B, and 20 in BI class C), and in the second phase, 40 patients were included (20 patients in BI class B and 20 patients in BI class C).
Of the patients, 94% (n=84) had a caregiver, of which 40% (n=34) had a son or a daughter, 32% (n=27) had a spouse, 17% (n=14) had other relatives, and 11% (n=9) had a professional caregiver. The mean value of functional capacity measured by the BI was 58.31 (SD 24.5). Concerning mobility aids, 58% (n=52) of patients did not require it, 20% (n=18) required a walker, 17% (n=15) a wheelchair, 4% (n=4) required a cane, and 1% (n=1) required crutches. The clinical complexity was high with 76 (85%) patients meeting the criteria for patients with polypathology, with a mean of 2.7 (SD 1.25), and 19 (21%) patients met the criteria for patients with complex chronic diseases. A total of 61 (69%) patients met the frailty criteria.
The most characteristic symptoms of this population were dyspnea (n=73, 82%), with 47% (n=42) of patients requiring home oxygen therapy; chronic pain (n=63, 70%); asthenia (n=61, 68%); and anxiety (n=41, 46%; Table 1 ). The mean number of drugs taken chronically was 12.19 (SD 11.88), with 87% (n=78) meeting the polypharmacy criteria and 70% (n=63) meeting the extreme polypharmacy criteria. Psychotropic drugs were the most consumed pharmacological group (n=29, 33%). Five patients died during the study.
a BI: Barthel Index.
b HRF: high risk of fall.
c PP: polypathological patient.
d PCCDs: patients with complex chronic diseases.
Regarding the baseline characteristics, comparing the different phases 1 and 2 categories, significant differences were only found in the presence of pain in the classification BI class B ( Table 1 ). In that category, the mean BI at the beginning of the study was 50.25 (SD 10.44), with an increase at the end of the study to 63.53 (SD 28.92). In BI class C, an initial BI of 80.75 (SD 11.27) and a final BI of 86.76 (SD 15.806) were observed. In the initial balance test for BI class B, the value was 1.6 (SD 1.46) points and 4.8 (SD 1.61) points at the end of the study, while in the BI class C, the initial value was 4 (SD 1.6) and the final value was 5.5 (SD 2.1).
Data for the ML Model
Following the aforementioned methodology, the patient cohort was reduced to 54 patients, taking into account only those who had at least 14 days’ worth of records before the doctor’s appointment. The principal factors contributing to this lack of data were predominantly attributed to mortality or patients becoming bedridden, subsequently ceasing to use the wristband. Mobility profiles corresponding to encounter 3 were used as a production-like test in a cohort of 21 patients. The best model results for defining the average 24-hour activity profile were obtained using the maximum value of steps in each interval and the type of walking aid if needed. Table 2 presents the complete set of variables used for training the model. The mean BI in the training and validation sets are 66.5 (SD 23.4) and 65.0 (SD 24.1), respectively. In contrast, the mean value in the test set is notably higher at 85.0 (SD 22.5).
The fitted model, whose parameters were selected through cross-validation, is a decision tree regressor with a depth of 3, minimum impurity decrease of 0.0, minimum samples in a leaf node of 2 and minimum samples in a split of 9 ( Figure 4 ). Among the features considered, the most important variables for predicting the BI were identified as the maximum step count during the evening and morning periods, and the absence of a mobility device. These key predictors were determined based on their significant impact on the functional status of the patients ( Figure 5 [ 28 ]).
Based on the results in Table 3 , the model exhibits consistency in MAD with a value close to 5 in the training, validation, and test sets. Furthermore, according to Figure 6 , when observing the predicted values compared to the real ones, the model does not present a significant difference between the predicted and the real BI.
Once the BI prediction was performed, the intervals defining each BI class were further considered. Subsequently, a classification prediction is carried out by converting the predicted value into its corresponding class label, thereby assigning the appropriate class to the given BI prediction. As observed in Table 3 and Figure 7 , in the training set, the model achieved precision, recall, and F 1 -scores of 0.88, 0.93, and 0.90 for class B, respectively. For class C, the model obtained precision, recall, and F 1 -scores of 0.94 for all 3 measures. However, for class A, all the metrics were 0.00 due to the limited support for that class (only 1 instance). In the validation set, the model demonstrated consistent performance with precision, recall, and F 1 -scores of 0.88 for both class B and class C. On the other hand, in the data coming from the test set, the model achieved precision, recall, and F 1 -scores of 0.5, 1, and 0.67 for class B, respectively. For class C, the model obtained precision, recall, and F 1 -scores of 1, 0.94, and 0.97, respectively. Finally, only 1 member from class A was predicted as class B.

a MAE: mean absolute error.
b MAD: median absolute error.
c RMSE: root mean squared error.

The use of mobility recording devices identifies different mobility patterns and relates them to functional capacity in patients with polypathology. One of this study’s findings is improving functional capacity measured by the BI in patients after using the mobility devices for 6 months in a real environment. This improvement is slight, 13 points in the case of moderate dependence and 6 points in mild dependence. We believe that it reflects an effect of the empowerment experienced by patients with the use of mobility devices. Therefore, the use of such mobility monitoring devices may have a potential impact on the management of complex chronic patients and could be included as part of clinical follow-up practices. Specifically, during the study execution in a real environment, patient empowerment was related to patient participation in decision-making, gaining control, and learning about their health.
In addition, patients’ sense of empowerment was related to less frustration with the technology [ 29 ]. This effect is well-known in the literature. A systematic review of 71 articles analyzing patients’ expectations of digital tools found that mobile apps increase patient engagement and motivation, especially when they can visualize parameters graphically and thus monitor their outcomes over time [ 30 ]. The evidence is scarce in patients with polypathology; but in another study of our group, we found similar results with slight improvements in functional capacity [ 31 ]. This empowerment is going to help patients in the self-management of their diseases. Health status has shown that incorporating digital technology into patients’ lives increases their awareness of lifestyle behaviors, which has helped them understand how to manage their health better and promote autonomy [ 32 ]. Longer term studies are needed to confirm this benefit, although it could be an alternative to integrate into the clinical practice of these patients to minimize their functional impairment.
Another possible beneficial effect of continuous monitoring of the functional capacity of patients with complex chronic diseases is the early detection of functional deterioration that may be the beginning of exacerbations of their diseases. If these data are integrated into the health care computer system, alarm situations could be determined that would allow early reaction by health staff to treat such exacerbation, prevent its progression, and minimize the functional deterioration that could be caused to the patient.
It should be noted that commercially available mobility monitoring devices have been used for this study and devices specifically designed for the study were not required. This favors cost reduction when considering the implementation of activity monitoring in patients with polypathology in real-world settings. Since the population with mobility devices is growing, with 515 million units sold in 2022, and patients with polypathology are a population that continue to increase and probably have their own mobility measurement device [ 33 ], the costs are thus reduced by integrating data in the informatics systems of the different health care organizations.
Another contribution of this study was to determine that mobility devices do not accurately recognize patients’ steps when using walking aids. For that reason, and to avoid this possible bias, patients were categorized into 3 groups depending on the walking aids. Additionally, caregivers assisting patients during physical activity have been classified similarly to canes or walkers due to their similarity in providing walking support. After the inclusion of an extra variable with the group to which the patient belongs, the ML model has managed to alleviate these limitations, achieving a good performance.
Concerning the data for the ML model, the use of the maximum value of steps taken in each of the 4 intervals defined (morning-afternoon-evening-overnight) yielded the most promising outcomes. This finding can be attributed to the limited and typically short-lived movements observed in patients with complex chronic conditions, which rarely extend beyond an hour. By leveraging the maximum step count within each time segment, we effectively capture the most significant and representative activity level during that period, thus optimizing the model’s performance. The selected intervals concur with the typical Spanish timetable for meals. Furthermore, the mean and SD data values found in the training and validation sets were similar, suggesting consistent levels of variability in both data sets. Moreover, including the heart rate information, measured by the WAT, and its relationship with the step count is proposed as a future study. Therefore, it could help to distinguish the requirement of the physical activity considering the cross-correlation or the cosine similarity between the step count and the heart rate.
Partitioning the data set into 7 equally sized subsets, with each fold serving as a validation set while the remaining folds are used for training, ensures robustness in selecting the optimal settings that yield the best predictive accuracy for the BI [ 34 ]. The hyperparameters play a crucial role in controlling the complexity and generalization of the decision tree model. By tuning these hyperparameters, the cross-validation process aims to find the optimal combination that balances model complexity and performance, resulting in a decision tree model with improved predictive capabilities [ 28 ].
Using a decision tree as a regression model holds paramount importance in the biomedicine field, particularly due to the necessity of using a highly interpretable model that can be effectively used and comprehended by the medical team [ 35 , 36 ]. The interpretability of the model enables medical professionals to understand the underlying decision-making process and gain insights into the factors influencing the predictions. This transparency fosters trust and facilitates collaboration between the model and the clinicians. Although more complex approaches exist, such as random forest or extreme gradient boosting, the ability to provide better results than decision trees in terms of accuracy most of the time, their lack of interpretability, and the limited sample size of this study advise against its use. Under these circumstances, a valid alternative to regression trees is multiple linear regression. However, a linear regression model based on the same variables as the decision tree has been performed, yielded inferior results.
Regarding the model performance, upon comparing the results obtained from the model’s predictions with the ground truth, the decision tree model generates accurate predictions. Therefore, the decision tree model can assess the functional capacity of patients based on data collected from the WAT. As observed, the errors remain similar among the training, validation, and test sets. Hence, this confirms that the model can generalize to unseen cases.
There is an imbalanced distribution of classes in the production-like test set, as shown in Figure 7 . This discrepancy arises from the natural transition of patients between classes B to C and A, due to changes in the BI, coupled with the criterion of minimum data required for data set inclusion. It is noteworthy that the test set could have been randomized to achieve an even distribution of class numbers, akin to the training and validation sets. However, given the primary goal of evaluating the model’s performance in a realistic production setting, this randomization was deliberately omitted.
In addition, it is worth noting that the MAD is the most suitable performance measure in this case. This choice is justified by the variability in step measurements captured by the WAT and the relatively small data set. It is possible that a patient with inaccurately measured data could significantly influence the error measure, particularly in terms of absolute or squared errors. By considering the MAD as the primary metric, we mitigate the impact of outliers or measurement inconsistencies, ensuring a more robust evaluation of the model’s performance in predicting the BI.
On the other hand, when considering the performance obtained in the classification problem, the accuracy remains consistent in training, validation, and test sets. The main objective of performing regression followed by classification into classes A, B, and C is due to the continuous nature of the BI variable. When aggregated into these 3 intervals, estimating BI solely through classification becomes complex, as small differences in BI values may result in a class label change. Therefore, regression allows the model to capture the underlying continuous relationship within the BI data, enhancing its ability to make more accurate and robust predictions while assigning the appropriate class labels based on the predicted values.
Furthermore, the model does not need to include clinical information such as specific disease, number of comorbidities, severity of disease, and so forth. Therefore, it can be regarded as a general model for patients with complex chronic diseases without specific clinical data, facilitating the development of a methodology that allows estimating the BI at any given moment using the information collected by the WAT over the last 30 days.
This study has several limitations. Since this was a pilot study with a small number of patients, the results should be confirmed by studies with a larger population. Prospective studies are needed to analyze whether identifying mobility changes and their transfer to health care systems can have care implications and improve the health status of patients with multiple pathologies. A notable element is that the bracelet does not register well the physical activity of patients who use a cane or crutches (or a wheelchair) since it cannot measure steps. A priori, this could be a limitation of the study. Still, adjusting the model by identifying walking aid devices and evaluating other parameters makes it possible to identify and predict mobility patterns in these patients.
In conclusion, using commercially available WATs makes it possible to identify different mobility patterns and relate them to functional capacity in patients with polypathology according to the BI without using clinical parameters.
Acknowledgments
This work was supported by the chronic-IoT project (Agencia Estatal de Investigación, PID2019- 110747RB-C21/AEI/10.13039/501100011033), which has received funding from the Ministry of Science, Innovation and Universities of the Government of Spain and the State Research Agency. Also, this research has been cosupported by the Carlos III National Institute of Health, through the IMPaCT-Data program (code IMP/00019), and through the Platform for Dynamization and Innovation of the Spanish National Health System industrial capacities and their effective transfer to the productive sector (code PT20/00088), both co-funded by European Regional Development Fund (FEDER) “A way of making Europe.”
Data Availability
The data sets generated during and analyzed during this study are not publicly available because there is no Ethics Committee approval for this purpose, since the received Ethics Committee’s approval was related to carrying out this study. However, they could be available from the corresponding author on reasonable request and this authorization shall then be requested.
Conflicts of Interest
None declared.
- Ferrer C, Orozco D, Román P, Carreras M, Gutiérrez R, Nuño R. Strategy for addressing chronicity in the National Health System. Ministerio de Sanidad, Servicios Sociales e Igualdad. 2021. URL: https://cpage.mpr.gob.es [accessed 2024-03-19]
- Casademont J, Francia E, Torres O. Age of patients admitted to internal medicine departments in Spain: a twenty years perspective [Article in Spanish]. Med Clin (Barc). 2012;138(7):289-292. [ CrossRef ] [ Medline ]
- Bernabeu-Wittel M, Alonso-Coello P, Rico-Blázquez M, Del Campo RR, Gómez SS, Vales EC. Development of clinical practice guidelines for patients with comorbidity and multiple diseases. Rev Clin Esp (Barc). 2014;214(6):328-335. [ CrossRef ] [ Medline ]
- Mahoney FI, Barthel DW. Functional evaluation: the Barthel Index. Md State Med J. 1965;14:61-65. [ Medline ]
- Montero-Odasso M, Schapira M, Soriano ER, Varela M, Kaplan R, Camera LA, et al. Gait velocity as a single predictor of adverse events in healthy seniors aged 75 years and older. J Gerontol A Biol Sci Med Sci. 2005;60(10):1304-1309. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Tinetti ME, Williams TF, Mayewski R. Fall risk index for elderly patients based on number of chronic disabilities. Am J Med. 1986;80(3):429-434. [ CrossRef ] [ Medline ]
- Podsiadlo D, Richardson S. The timed "up and go": a test of basic functional mobility for frail elderly persons. J Am Geriatr Soc. 1991;39(2):142-148. [ CrossRef ] [ Medline ]
- Silguero SAA, Martínez-Reig M, Arnedo LG, Martínez GJ, Rizos LR, Soler PA. Chronic disease, mortality and disability in an elderly Spanish population: the FRADEA study [Article in Spanish]. Rev Esp Geriatr Gerontol. 2014;49(2):51-58. [ CrossRef ] [ Medline ]
- Cech DJ, Martin ST. Functional Movement Development Across the Life Span, 2nd Edition. St. Louis. Elsevier Health Sciences; 2011.
- Cid-Ruzafa J, Damián-Moreno J. Disability evaluation: Barthel's Index [Article in Spanish]. Rev Esp Salud Publica. 1997;71(2):127-137. [ FREE Full text ] [ Medline ]
- Caspersen CJ, Powell KE, Christenson GM. Physical activity, exercise, and physical fitness: definitions and distinctions for health-related research. Public Health Rep. 1985;100(2):126-131. [ FREE Full text ] [ Medline ]
- Booth FW, Roberts CK, Thyfault JP, Ruegsegger GN, Toedebusch RG. Role of inactivity in chronic diseases: evolutionary insight and pathophysiological mechanisms. Physiol Rev. 2017;97(4):1351-1402. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Sanz-Bobi MA, Contreras D, Sánchez Á. Multi-agent systems orientated to assist with daily activities in the homes of elderly and disabled people. In: Zacarias M, de Oliveira JV, editors. Human-Computer Interaction: The Agency Perspective. Studies in Computational Intelligence, Vol 396. Berlin, Heidelberg. Springer; 2012;131-167.
- Ohta S, Nakamoto H, Shinagawa Y, Tanikawa T. A health monitoring system for elderly people living alone. J Telemed Telecare. 2002;8(3):151-156. [ CrossRef ] [ Medline ]
- Lyons GM, Culhane KM, Hilton D, Grace PA, Lyons D. A description of an accelerometer-based mobility monitoring technique. Med Eng Phys. 2005;27(6):497-504. [ CrossRef ] [ Medline ]
- Velasco MA, López-Blanco R, Serrano JI, del Castillo MD, Romero JP, Benito-León J, et al. Design of a platform; HEALTH based on smart watches for home monitoring of neurological diseases: NETMD. Cogn Area Networks. 2024.:31.
- Asada HH, Shaltis P, Reisner A, Rhee S, Hutchinson RC. Mobile monitoring with wearable photoplethysmographic biosensors. IEEE Eng Med Biol Mag. 2003;22(3):28-40. [ CrossRef ] [ Medline ]
- Pantelopoulos A, Bourbakis NG. A survey on wearable sensor-based systems for health monitoring and prognosis. IEEE Trans Syst Man Cybern C. 2010;40(1):1-12. [ CrossRef ]
- Bhelkar V, Shedge DK. Different types of wearable sensors and health monitoring systems: a survey. 2016. Presented at: 2016 2nd International Conference on Applied and Theoretical Computing and Communication Technology (iCATccT); July 21-23, 2016;43-48; Bangalore, India. [ CrossRef ]
- Olson JS, Redkar S. A survey of wearable sensor networks in health and entertainment. MOJ Appl Bionics Biomech. 2018;2(5):280-287. [ FREE Full text ] [ CrossRef ]
- Louter M, Maetzler W, Prinzen J, van Lummel RC, Hobert M, Arends JBAM, et al. Accelerometer-based quantitative analysis of axial nocturnal movements differentiates patients with Parkinson's disease, but not high-risk individuals, from controls. J Neurol Neurosurg Psychiatry. 2015;86(1):32-37. [ CrossRef ] [ Medline ]
- Tucker CS, Behoora I, Nembhard HB, Lewis M, Sterling NW, Huang X. Machine learning classification of medication adherence in patients with movement disorders using non-wearable sensors. Comput Biol Med. 2015;66:120-134. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Kobsar D, Ferber R. Wearable sensor data to track subject-specific movement patterns related to clinical outcomes using a machine learning approach. Sensors (Basel). 2018;18(9):2828. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Jalloul N. Wearable sensors for the monitoring of movement disorders. Biomed J. 2018;41(4):249-253. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Polo-Molina A, Sánchez-Úbeda EF, Portela J, Palacios R, Rodríguez-Morcillo C, Muñoz A, et al. Analyzing mobility patterns of complex chronic patients using wearable activity trackers: a machine learning approach. Eng Proc. 2023;39(1):92. [ CrossRef ]
- Ollero MB, Bernabeu-Wittel M, Almendro E, Manuel J, Raúl GE, Herrera M, et al. Integrated Care Process. Care for Multipathological Patients, 3rd Edition. Andalucia. Consejería de Salud; 2018.
- Shah S, Vanclay F, Cooper B. Improving the sensitivity of the Barthel index for stroke rehabilitation. J Clin Epidemiol. 1989;42(8):703-709. [ CrossRef ] [ Medline ]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825-2830. [ FREE Full text ]
- Risling T, Martinez J, Young J, Thorp-Froslie N. Evaluating patient empowerment in association with eHealth technology: scoping review. J Med Internet Res. 2017;19(9):e329. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Madanian S, Nakarada-Kordic I, Reay S, Chetty T. Patients' perspectives on digital health tools. PEC Innov. 2023;2:100171. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Hernandez-Quiles C, Bernabeu-Wittel M, Barón-Franco B, Palacios AA, Garcia-Serrano MR, Lopez-Jimeno W, et al. A randomized clinical trial of home telemonitoring in patients with advanced heart and lung diseases. J Telemed Telecare. 2024;30(2):356-364. [ CrossRef ] [ Medline ]
- Du Y, Dennis B, Rhodes SL, Sia M, Ko J, Jiwani R, et al. Technology-assisted self-monitoring of lifestyle behaviors and health indicators in diabetes: qualitative study. JMIR Diabetes. 2020;5(3):e21183. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Wearables market in 2022. News Europe. URL: https://www.eenewseurope.com/en/wearables-market-fell-in-2022-says-idc/ [accessed 2023-06-13]
- James G, Witten D, Hastie T, Tibshirani R. An Introduction to Statistical Learning: With Applications in R. New York. Springer; 2013.
- Elshawi R, Al-Mallah MH, Sakr S. On the interpretability of machine learning-based model for predicting hypertension. BMC Med Inform Decis Mak. 2019;19(1):146. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Caruana R, Lou Y, Gehrke J, Koch P, Sturm M, Elhadad N. Intelligible models for healthcare: predicting pneumonia risk and hospital 30-day readmission. 2015. Presented at: KDD '15: Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; August 10-13, 2015;1721-1730; Sydney, NSW, Australia. [ CrossRef ]
Abbreviations
Edited by A Mavragani; submitted 18.09.23; peer-reviewed by S Okita; comments to author 03.01.24; revised version received 18.01.24; accepted 19.02.24; published 19.04.24.
©Celia Alvarez-Romero, Alejandro Polo-Molina, Eugenio Francisco Sánchez-Úbeda, Carlos Jimenez-De-Juan, Maria Pastora Cuadri-Benitez, Jose Antonio Rivas-Gonzalez, Jose Portela, Rafael Palacios, Carlos Rodriguez-Morcillo, Antonio Muñoz, Carlos Luis Parra-Calderon, Maria Dolores Nieto-Martin, Manuel Ollero-Baturone, Carlos Hernández-Quiles. Originally published in JMIR Formative Research (https://formative.jmir.org), 19.04.2024.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work, first published in JMIR Formative Research, is properly cited. The complete bibliographic information, a link to the original publication on https://formative.jmir.org, as well as this copyright and license information must be included.
A Review on Analyzing and Predicting the State of Cancer Disease using Machine Learning Algorithms
Ieee account.
- Change Username/Password
- Update Address
Purchase Details
- Payment Options
- Order History
- View Purchased Documents
Profile Information
- Communications Preferences
- Profession and Education
- Technical Interests
- US & Canada: +1 800 678 4333
- Worldwide: +1 732 981 0060
- Contact & Support
- About IEEE Xplore
- Accessibility
- Terms of Use
- Nondiscrimination Policy
- Privacy & Opting Out of Cookies
A not-for-profit organization, IEEE is the world's largest technical professional organization dedicated to advancing technology for the benefit of humanity. © Copyright 2024 IEEE - All rights reserved. Use of this web site signifies your agreement to the terms and conditions.

IMAGES
VIDEO
COMMENTS
Disease Prediction Using Machine Learning. * Research Gate Link: Marouane Fethi Ferjani. Computing Department. Bournemouth University. Bournemouth, England. [email protected]. Abstract ...
Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis: 257 : Random forest-based similarity measures for multi-modal classification of Alzheimer's disease: 248 : Effective Heart disease prediction Using hybrid Machine Learning techniques: 214
Developing a medical diagnosis system based on machine learning (ML) algorithms for prediction of any disease can help in a more accurate diagnosis than the conventional method. We have designed a disease prediction system using multiple ML algorithms. The data set used had more than 230 diseases for processing.
Study characteristics. Table 2 shows the basic characteristics of the included studies. In total, our meta-analysis of ML and cardiovascular diseases included 103 cohorts (55 studies) with a total ...
ChaoTan et al [1] explored the feasibility of using decision stumps as a poor classification method and track element analysis to predict timely lung cancer in a combination of Adaboost (machine learning ensemble). For the illustration, a cancer dataset was used which identified 9 trace elements in 122 urine samples.
The numbers of disease prediction papers using XGBoost with medical data have increased recently 33,34,35,36. XGBoost is an algorithm that overcomes the shortcomings of GBM (gradient boosting ...
Using machine learning and deep learning platforms, this study aims to combine recent research on four brain diseases: Alzheimer's disease, brain tumors, epilepsy, and Parkinson's disease. By using 22 brain disease databases that are used most during the reviews, the authors can determine the most accurate diagnostic method.
6 Conclusion. This paper reviews the deep learning algorithms in the field of disease prediction. According to the type of data processed, the algorithms are divided into structured data algorithms and unstructured data algorithms. Structured data algorithms include ANN and FM-Deep Learning algorithms.
Comparatively, supervised machine learning (ML) algorithms has shown notable capability in exceeding standard approach for disease detection and helps medical experts in the early detection of high-risk diseases. In this paper, algorithms discussed were K- Nearest Neighbor, Naïve Bayes, Support Vector Machine and Decision Trees.
Background Supervised machine learning algorithms have been a dominant method in the data mining field. Disease prediction using health data has recently shown a potential application area for these methods. This study aims to identify the key trends among different types of supervised machine learning algorithms, and their performance and usage for disease risk prediction. Methods In this ...
Purpose Disease risk prediction poses a significant and growing challenge in the medical field. While researchers have increasingly utilised machine learning (ML) algorithms to tackle this issue, supervised ML methods remain dominant. However, there is a rising interest in unsupervised techniques, especially in situations where data labels might be missing — as seen with undiagnosed or rare ...
Disease Prediction using Machine Learning Abstract: The dependency on computer-based technology has resulted in storage of lot of electronic data in the health care industry. As a result of which, health professionals and doctors are dealing with demanding situations to research signs and symptoms correctly and perceive illnesses at an early stage.
In this research, the potential of utilizing media data to predict if an infectious disease will break out or not in a particular country using three of the most widely used machine learning ...
This research work carried out demonstrates the disease prediction system developed using Machine learning algorithms such as Decision Tree classifier, Random forest classifier, and Naïve Bayes classifier. The paper presents the comparative study of the results of the above algorithms used.
In this research paper, we have applied three machine learning classifiers logistic regression, decision tree and support vector machine on chronic kidney diseases dataset collected from UCI machine learning repository. ... Pal, S. Chronic Kidney Disease Prediction Using Machine Learning Techniques. Biomedical Materials & Devices 1, 534-540 ...
The COVID-19 pandemic has presented unprecedented public health challenges worldwide. Understanding the factors contributing to COVID-19 mortality is critical for effective management and intervention strategies. This study aims to unlock the predictive power of data collected from personal, clinical, preclinical, and laboratory variables through machine learning (ML) analyses.
This approach makes use of a dataset from the UCI Machine Learning Repository 11 referred to as CKD. A total of 24 features and 1 target variable are included in the CKD Dataset. It can be broken down into 2 categories, yes or no. The dataset has 25 attributes, 11 of which are numerical and 14 of which are nominal.
Machine learning models are used to create and enhance various disease prediction frameworks. Ensemble learning is a machine learning technique that combines multiple classifiers to improve performance by making more accurate predictions than a single classifier. Although numerous studies have employed ensemble approaches for disease prediction, there is a lack of thorough assessment of ...
This paper is an exploration towards the applications of machine learning techniques in the context of multiple disease prediction, which aims towards the enhancement of diagnostic accuracy and facilitates timely invention. ... Pooja and Patil, Mohini and Suryawanshi, Gayatri and Chaphadkar, Anagha, Multiple Disease Prediction using Machine ...
Section 2 of this paper will introduce the theories, development and disease application cases of two kinds of structured data algorithms, ANN and FM-Deep Learning. Section 3 will introduce the theories, development and disease application cases of CNN and RNN. Section 4 will respectively introduce the current defects in the field of disease prediction algorithms and the coping strategies.
"Multiple Disease Prediction Using Machine Learning Algorithms" by Chauhan et al. (2021): This paper investigates using various ML algorithms, including SVM and Decision Trees, for multiple disease ... Exploring how to enhance accessibility for healthcare practitioners and ensuring ease of use could be a valuable research focus. 5.
Cardiovascular disease refers to any critical condition that impacts the heart. Because heart diseases can be life-threatening, researchers are focusing on designing smart systems to accurately diagnose them based on electronic health data, with the aid of machine learning algorithms. This work presents several machine learning approaches for predicting heart diseases, using data of major ...
This paper proposed a method of identification and prediction of the presence of chronic disease in an individual using the machine learning algorithms such as CNN and KNN. The advantage of the proposed system is the use of both structured and unstructured data from real life for data set preparation, which lacks in many of the existing approaches.
Statistical analysis was performed using SPSS and Python for the machine learning modeling. Results: Overall, 90 patients with complex chronic diseases were included: 50 during phase 1 (class A: n=10; class B: n=20; and class C: n=20) and 40 during phase 2 (class B: n=20 and class C: n=20). Most patients (n=85, 94%) had a caregiver.
The main aim of this study is to evaluate various existing Machine Learning and optimization approaches, identifying the most suitable methods to accommodate extensive datasets with high prediction accuracy. This revision primarily aims to showcase previous research on machine-learning techniques employed for cancer detection.
The "Symptoms-Based Disease Prediction System using Machine Learning" project is a solution that intends to help users predict their potential diseases based on the symptoms they enter and other general information, which is implemented using the Python programming language. Abstract: The "Symptoms-Based Disease Prediction System using Machine Learning" project is a solution that intends to ...