
- Deep Learning Research Proposal
The word deep learning is the study and analysis of deep features that are hidden in the data using some intelligent deep learning models . Recently, it turns out to be the most important research paradigm for advanced automated systems for decision-making . Deep learning is derived from machine learning technologies that learn based on hierarchical concepts . So, it is best for performing complex and long mathematical computations in deep learning .
This page describes to you the innovations of deep learning research proposals with major challenges, techniques, limitations, tools, etc.!!!
One most important thing about deep learning is the multi-layered approach . It enables the machine to construct and work the algorithms in different layers for deep analysis . Further, it also works on the principle of artificial neural networks which functions in the same human brain. Since it got inspiration from the human brain to make machines automatically understand the situation and make smart decisions accordingly. Here, we have given you some of the important real-time applications of deep learning.

Deep Learning Project Ideas
- Natural Language Processing
- Pattern detection in Human Face
- Image Recognition and Object Detection
- Driverless UAV Control Systems
- Prediction of Weather Condition Variation
- Machine Translation for Autonomous Cars
- Medical Disorder Diagnosis and Treatment
- Traffic and Speed Control in Motorized Systems
- Voice Assistance for Dense Areas Navigation
- Altitude Control System for UAV and Satellites
Now, we can see the workflow of deep learning models . Here, we have given you the steps involved in the deep learning model. This assists you to know the general procedure of deep learning model execution . Similarly, we precisely guide you in every step of your proposed deep learning model . Further, the steps may vary based on the requirement of the handpicked deep learning project idea. Anyway, the deep learning model is intended to grab deep features of data by processing through neural networks . Then, the machine will learn and understand the sudden scenarios for controlling systems.

Process Flow of Deep Learning
- Step 1 – Load the dataset as input
- Step 2 – Extraction of features
- Step 3 – Process add-on layers for more abstract features
- Step 4 – Perform feature mapping
- Step 5 –Display the output
Although deep learning is more efficient to automatically learn features than conventional methods, it has some technical constraints. Here, we have specified only a few constraints to make you aware of current research. Beyond these primary constraints, we also handpicked more number of other constraints. To know other exciting research limitations in deep learning , approach us. We will make you understand more from top research areas.
Deep Learning Limitations
- Test Data Variation – When the test data is different from training data, then the employed deep learning technique may get failure. Further, it also does not efficiently work in a controlled environment.
- Huge Dataset – Deep learning models efficiently work on large-scale datasets than limited data
Our research team is highly proficient to handle different deep learning technologies . To present you with up-to-date information, we constantly upgrade our research knowledge in all advanced developments. So, we are good not only at handpicking research challenges but also more skilled to develop novel solutions. For your information, here we have given you some most common data handling issues with appropriate solutions.
What are the data handling techniques?
- Variables signifies the linear combo of factors with errors
- Depends on the presence of different unobserved variables (i.e., assumption)
- Identify the correlations between existing observed variables
- If the data in a column has fixed values, then it has “0” variance.
- Further, these kinds of variables are not considered in target variables
- If there is the issue of outliers, variables, and missing values, then effective feature selection will help you to get rid out of it.
- So, we can employ the random forest method
- Remove the unwanted features from the model
- Repeat the same process until attaining maximum error rate
- At last, define the minimum features
- Remove one at a time and check the error rate
- If there are dependent values among data columns, then may have redundant information due to similarities.
- So, we can filter the largely correlated columns based on coefficients of correlation
- Add one at a time for high performance
- Enhance the entire model efficiency
- Addresses the possibility where data points are associated with high-dimensional space
- Select low-dimensional embedding to generate related distribution
- Identify the missing value columns and remove them by threshold
- Present variable set is converted to a new variable set
- Also, referred to as a linear combo of new variables
- Determine the location of each point by pair-wise spaces among all points which are represented in a matrix
- Further, use standard multi-dimensional scaling (MDS) for determining low-dimensional points locations
In addition, we have also given you the broadly utilized deep learning models in current research . Here, we have classified the models into two major classifications such as discriminant models and generative models . Further, we have also specified the deep learning process with suitable techniques. If there is a complex situation, then we design new algorithms based on the project’s needs . On the whole, we find apt solutions for any sort of problem through our smart approach to problems.
Deep Learning Models
- CNN and NLP (Hybrid)
- Domain-specific
- Image conversion
- Meta-Learning
Furthermore, our developers are like to share the globally suggested deep learning software and tools . In truth, we have thorough practice on all these developing technologies. So, we are ready to fine-tuned guidance on deep learning libraries, modules, packages, toolboxes , etc. to ease your development process. By the by, we will also suggest you best-fitting software/tool for your project . We ensure you that our suggested software/tool will make your implementation process of deep learning projects techniques more simple and reliable .
Deep Learning Software and Tools
- Caffe & Caffe2
- Deep Learning 4j
- Microsoft Cognitive Toolkit
So far, we have discussed important research updates of deep learning . Now, we can see the importance of handpicking a good research topic for an impressive deep learning research proposal. In the research topic, we have to outline your research by mentioning the research problem and efficient solutions . Also, it is necessary to check the future scope of research for that particular topic.
The topic without future research direction is not meant to do research!!!
For more clarity, here we have given you a few significant tips to select a good deep learning research topic.
How to write a research paper on deep learning?
- Check whether your selected research problem is inspiring to overcome but not take more complex to solve
- Check whether your selected problem not only inspires you but also create interest among readers and followers
- Check whether your proposed research create a contribution to social developments
- Check whether your selected research problem is unique
From the above list, you can get an idea about what exactly a good research topic is. Now, we can see how a good research topic is identified.
- To recognize the best research topic, first undergo in-depth research on recent deep learning studied by referring latest reputed journal papers.
- Then, perform a review process over the collected papers to detect what are the current research limitations, which aspect not addressed yet, which is a problem is not solved effectively, which solution is needed to improve, what the techniques are followed in recent research, etc.
- This literature review process needs more time and effort to grasp knowledge on research demands among scholars.
- If you are new to this field, then it is suggested to take the advice of field experts who recommend good and resourceful research papers.
- Majorly, the drawbacks of the existing research are proposed as a problem to provide suitable research solutions.
- Usually, it is good to work on resource-filled research areas than areas that have limited reference.
- When you find the desired research idea, then immediately check the originality of the idea. Make sure that no one is already proved your research idea.
- Since, it is better to find it in the initial stage itself to choose some other one.
- For that, the search keyword is more important because someone may already conduct the same research in a different name. So, concentrate on choosing keywords for the literature study.
How to describe your research topic?
One common error faced by beginners in research topic selection is a misunderstanding. Some researchers think topic selection means is just the title of your project. But it is not like that, you have to give detailed information about your research work on a short and crisp topic . In other words, the research topic is needed to act as an outline for your research work.
For instance: “deep learning for disease detection” is not the topic with clear information. In this, you can mention the details like type of deep learning technique, type of image and its process, type of human parts, symptoms , etc.
The modified research topic for “deep learning for disease detection” is “COVID-19 detection using automated deep learning algorithm”
For your awareness, here we have given you some key points that need to focus on while framing research topics. To clearly define your research topic, we recommend writing some text explaining:
- Research title
- Previous research constraints
- Importance of the problem that overcomes in proposed research
- Reason of challenges in the research problem
- Outline of problem-solving possibility
To the end, now we can see different research perspectives of deep learning among the research community. In the following, we have presented you with the most demanded research topics in deep learning such as image denoising, moving object detection, and event recognition . In addition to this list, we also have a repository of recent deep learning research proposal topics, machine learning thesis topics . So, communicate with us to know the advanced research ideas of deep learning.
Research Topics in Deep Learning
- Continuous Network Monitoring and Pipeline Representation in Temporal Segment Networks
- Dynamic Image Networks and Semantic Image Networks
- Advance Non-uniform denoising verification based on FFDNet and DnCNN
- Efficient image denoising based on ResNets and CNNs
- Accurate object recognition in deep architecture using ResNeXts, Inception Nets and Squeeze and Excitation Networks
- Improved object detection using Faster R-CNN, YOLO, Fast R-CNN, and Mask-RCNN

Overall, we are ready to support you in all significant and new research areas of deep learning . We guarantee you that we provide you novel deep learning research proposal in your interested area with writing support. Further, we also give you code development , paper writing, paper publication, and thesis writing services . So, create a bond with us to create a strong foundation for your research career in the deep learning field.
Related Pages
Services we offer.
Mathematical proof
Pseudo code
Conference Paper
Research Proposal
System Design
Literature Survey
Data Collection
Thesis Writing
Data Analysis
Rough Draft
Paper Collection
Code and Programs
Paper Writing
Course Work
reason.town

How to Write a Machine Learning Research Proposal
Introduction, what is a machine learning research proposal, the structure of a machine learning research proposal, tips for writing a machine learning research proposal, how to get started with writing a machine learning research proposal, the importance of a machine learning research proposal, why you should take the time to write a machine learning research proposal, how to make your machine learning research proposal stand out, the bottom line: why writing a machine learning research proposal is worth it, further resources on writing machine learning research proposals.
If you want to get into machine learning, you first need to get past the research proposal stage. We’ll show you how.
Checkout this video:

A machine learning research proposal is a document that summarizes your research project, methods, and expected outcomes. It is typically used to secure funding for your project from a sponsor or institution, and can also be used to assessment your project by peers. Your proposal should be clear, concise, and well-organized. It should also provide enough detail to allow reviewers to assess your project’s feasibility and potential impact.
In this guide, we will cover the basics of what you need to include in a machine learning research proposal. We will also provide some tips on how to create a strong proposal that is more likely to be funded.
A machine learning research proposal is a document that describes a proposed research project that uses machine learning algorithms and techniques. The proposal should include a brief overview of the problem to be tackled, the proposed solution, and the expected results. It should also briefly describe the dataset to be used, the evaluation metric, and any other relevant details.
There is no one-size-fits-all answer to this question, as the structure of a machine learning research proposal will vary depending on the specific research question you are proposing to answer, the methods you plan to use, and the overall focus of your proposal. However, there are some general principles that all good proposals should follow.
In general, a machine learning research proposal should include:
-A summary of the problem you are trying to solve and the motivation for solving it -A brief overview of previous work in this area, including any relevant background information -A description of your proposed solution and a discussion of how it compares to existing approaches -An evaluation plan outlining how you will evaluate the effectiveness of your proposed solution -A discussion of any potential risks or limitations associated with your proposed research
Useful tips for writing a machine learning research proposal:
-Your proposal should address a specific problem or question in machine learning.
-Before writing your proposal, familiarize yourself with the existing literature in the field. Your proposal should build on the existing body of knowledge and contribute to the understanding of the chosen problem or question.
-Your proposal should be clear and concise. It should be easy for non-experts to understand what you are proposing and why it is important.
-Your proposal should be well organized. Include a brief introduction, literature review, methodology, expected results, and significance of your work.
-Make sure to proofread your proposal carefully before submitting it.
A machine learning research proposal is a document that outlines the problem you want to solve with machine learning, the methods you will use to solve it, the data you will use, and the anticipated results. This guide provides an overview of what should be included in a machine learning research proposal so that you can get started on writing your own.
1. Introduction 2. Problem statement 3. Methodology 4. Data 5. Evaluation 6. References
A machine learning research proposal is a document that outlines the rationale for a proposed machine learning research project. The proposal should convince potential supervisors or funding bodies that the project is worthwhile and that the researcher is competent to undertake it.
The proposal should include:
– A clear statement of the problem to be addressed or the question to be answered – A review of relevant literature – An outline of the proposed research methodology – A discussion of the expected outcome of the research – A realistic timeline for completing the project
A machine learning research proposal is not just a formal exercise; it is an opportunity to sell your idea to potential supervisors or funding bodies. Take advantage of this opportunity by doing your best to make your proposal as clear, concise, and convincing as possible.
Your machine learning research proposal is your chance to sell your project to potential supervisors and funders. It should be clear, concise and make a strong case for why your project is worth undertaking.
A well-written proposal will convince others that you have a worthwhile project and that you have the necessary skills and experience to complete it successfully. It will also help you to clarify your own ideas and focus your research.
Writing a machine learning research proposal can seem like a daunting task, but it doesn’t have to be. If you take it one step at a time, you’ll be well on your way to writing a strong proposal that will get the support you need.
In order to make your machine learning research proposal stand out, you will need to do several things. First, make sure that your proposal is well written and free of grammatical errors. Second, make sure that your proposal is clear and concise. Third, make sure that your proposal is organized and includes all of the necessary information. Finally, be sure to proofread your proposal carefully before submitting it.
The benefits of writing a machine learning research proposal go beyond helping you get funding for your project. A good proposal will also force you to think carefully about your problem and how you plan to solve it. This process can help you identify potential flaws in your approach and make sure that your project is as strong as possible before you start.
It can also be helpful to have a machine learning research proposal on hand when you’re talking to potential collaborators or presenting your work to a wider audience. A well-written proposal can give people a clear sense of what your project is about and why it’s important, which can make it easier to get buy-in and find people who are excited to work with you.
In short, writing a machine learning research proposal is a valuable exercise that can help you hone your ideas and make sure that your project is as strong as possible before you start.
Here are some further resources on writing machine learning research proposals:
– How to Write a Machine Learning Research Paper: https://MachineLearningMastery.com/how-to-write-a-machine-learning-research-paper/
– 10 Tips for Writing a Machine Learning Research Paper: https://blog.MachineLearning.net/10-tips-for-writing-a-machine-learning-research-paper/
Please also see our other blog post on writing research proposals: https://www.MachineLearningMastery.com/how-to-write-a-research-proposal/
Similar Posts

Scoring and Ranking in Machine Learning: What You Need to Know
ContentsIntroductionWhat is scoring and ranking in machine learning?Why is scoring and ranking important in machine learning?How is scoring and ranking used in machine learning?What are some benefits of scoring and ranking in machine learning?What are some challenges of scoring and ranking in machine learning?How can you improve your scoring and ranking in machine learning?ConclusionResourcesAbout the…

How to Get Machine Learning Grants
ContentsIntroductionWhat is Machine Learning?What are the benefits of Machine Learning?What are the types of Machine Learning?What are the applications of Machine Learning?How can Machine Learning be used to get grants?What are the steps involved in getting Machine Learning grants?What are the benefits of getting Machine Learning grants?What are the risks involved in getting Machine Learning…

What is Computer Vision?
ContentsWhat is Computer Vision?What are the goals of Computer Vision?What are the applications of Computer Vision?What are the challenges of Computer Vision?What is the history of Computer Vision?What are the state-of-the-art methods in Computer Vision?What are the datasets used in Computer Vision?What are the evaluation metrics used in Computer Vision?What are the tools used in…

Machine Learning and Your Credit Card
ContentsHow machine learning is changing the credit card industryThe benefits of using machine learning for credit card companiesThe benefits of using machine learning for consumersHow machine learning can help prevent credit card fraudThe future of machine learning in the credit card industryThe challenges of using machine learning for credit card companiesThe challenges of using machine…

Is C++ Good for Machine Learning?
ContentsIntroductionWhat is C++?What is Machine Learning?The Benefits of C++ for Machine LearningThe Drawbacks of C++ for Machine LearningThe Future of C++ and Machine LearningConclusionReferences If you’re wondering if C++ is a good language for machine learning, the answer is yes! Check out our blog post to learn more about the benefits of using C++ for…

Probability and Statistics for Machine Learning
ContentsIntroduction to Probability and StatisticsProbability FundamentalsStatistical InferenceRegression AnalysisClassification MethodsAnomaly DetectionDimensionality ReductionModel SelectionBoosting MethodsDeep Learning This blog post covers the basics of probability and statistics for machine learning. It covers topics such as probability distributions, statistical inference, and model selection. After reading this post, you will have a better understanding of how to apply these concepts…
EXPANSE, A Continual Deep Learning System; Research Proposal
Ieee account.
- Change Username/Password
- Update Address
Purchase Details
- Payment Options
- Order History
- View Purchased Documents
Profile Information
- Communications Preferences
- Profession and Education
- Technical Interests
- US & Canada: +1 800 678 4333
- Worldwide: +1 732 981 0060
- Contact & Support
- About IEEE Xplore
- Accessibility
- Terms of Use
- Nondiscrimination Policy
- Privacy & Opting Out of Cookies
A not-for-profit organization, IEEE is the world's largest technical professional organization dedicated to advancing technology for the benefit of humanity. © Copyright 2024 IEEE - All rights reserved. Use of this web site signifies your agreement to the terms and conditions.
Deep-learning Enhanced Healthcare Modeling and Optimization - Research - College of Engineering - Purdue University

Deep-learning Enhanced Healthcare Modeling and Optimization
Project Description
In the age of big data analytics, one must consider the continuum from predictive to prescriptive analytics to help managers to improve their day-to-day operations in large-scale healthcare systems. These systems often run under uncertainties and in rapidly changing environments. Good prescriptive management solutions require building high-fidelity models that are adaptive to the changing environment. Consequently, a framework for learning stochastic models from data in this setting is imperative. These learnt models need to be seamlessly integrated with data-driven prescriptive methods to optimize system operations.
In this research project, we work closely with the largest hospital systems in the state of Indiana and propose a methodological framework that collaboratively leverages deep learning and stochastic process theory to revolutionize workload prediction and resource planning, such as capacity and staffing. These developments are expected to enable fundamental improvements in short-term and long-term operations for healthcare delivery. Our research agenda, in support of this broader goal, includes (i) a novel framework for inferring stochastic models of time-varying, large-scale healthcare systems; (ii) a robust resource allocation framework that accounts for model uncertainty and natural stochastic variation; (iii) integration and deployment of our algorithms into all 16 hospitals belonging to the collaborating healthcare system.
Postdoc Qualifications
Applicants hold (or are about to complete) a PhD in Operations Research, Industrial Engineering, Applied Mathematics, Electrical Engineering or a related discipline. A strong background in stochastic modeling and optimization methods is required. Research experience in statistics/machine learning/deep learning would be a great advantage. A willingness to learn the fundamental theory and methods of statistics/machine learning/deep learning is necessary. Programming skills, fluency in English and excellent communication and presentation skills are essential.
Co-Advisors
Harsha Honnappa, [email protected], School of Industrial Engineering, engineering.purdue.edu/SSL
Pengyi Shi, [email protected], Krannert School of Business, https://web.ics.purdue.edu/~shi178
“Estimating Stochastic Poisson Intensities Using Deep Latent Models”, R. Wang, P. Jaiswal and H. Honnappa, Proceedings of the Winter Simulation Conference (2020).
“Timing it Right: Balancing Inpatient Congestion versus Readmission Risk at Discharge," P. Shi, J. E. Helm, J. Deglise-Hawkinson, and J. Pan. Operations Research, forthcoming
“The ∆(i)/GI/1 Queueing Model, and its Fluid and Diffusion Approximations”, H. Honnappa , R. Jain and A. R. Ward, Queueing Systems: Theory and Applications, 80.1-2 (2015): 71-103.
"Inpatient Bed Overflow: An Approximate Dynamic Programming Approach," J. G. Dai, P. Shi I Manufacturing and Service Operations Management. 2019; 21(4):894-911.
RESEARCH PROPOSAL DEEP LEARNING
Get your deep learning proposal work from high end trained professionals. The passion of your areas of interest will be clearly reflected in your proposal. Chose an expert to provide you with custom research proposal work. To interpret the real-time process of the art, historical context and future scopes we have made a literature survey in Deep Learning (DL).
- Define Objectives:
- Clearly sketch what we need to execute with our comprehensive view.
- Take transformers in Natural Language Processing (NLP) as an example and note its specific tasks and issues.
- Primary Sources:
- Research Databases: We can use the fields such as Google Scholar, arXIv, PubMed (for biomedical papers), IEEE Xplore, and others.
- Conference: Here NeurIPS, ICML, ICLR, CVPR, ICCV, ACL, EMNLP are the basic conferences in DL.
- Journal: The Journal of Machine Learning Research (JMLR) and Neural Computation are the papers frequently establish DL related studies.
- Start by Reviews and Surveys:
- Find the latest survey and review papers on our area of interest which gives a literature outline and frequently see the seminal latest works.
- Begin with Convolutional Neural Networks (CNNs) architecture survey paper if we search for CNN.
- Reading Papers:
- Skim: Begin with reading abstracts, introductions, conclusions, and figures.
- Deep Dive: When a study shows high similar to our work, then look in-depth to its methodology, experiments, and results.
- Take Notes: Look down the basic plans, methods, datasets, Evaluation metrics, and open issues described in the paper and note it.
- Forward and Backward Search:
- Forward: We can detect how the area is emerging using the tools such as Google Scholar’s “Cited by” feature to find latest papers in our research.
- Backward : We can track the improvement of designs by seeing the reference which is gives more knowledge in our study.
- Organize and Combine:
- Classify the papers by its themes, methodologies and version.
- We have to analyze the trends, patterns, and gaps in the literature.
- Keep Updates:
- We need to stay update with notifications on fields such as Google Scholar and arXiv for keywords similar to our title with the recent publications, because Dl is a fast-emerging area.
- Tools and Platforms:
- Utilize the tools such as Mendeley, Zotero and EndNote for maintaining and citing papers.
- We find similar papers with AI-driven suggestions from Semantic Scholar platform.
- Engage with the Community:
- Join into mailing lists, social media groups and online conference to get related with DL. Websites such as Reddit’s r/Machine Learning or the AI Alignment Forum frequently gather latest papers.
- By attending the webinars, workshops and meetings often can help us to gain skills from recent techniques and find knowledge of what the group seems essential.
- Report and Share:
- If we want to establish the paper make annotated bibliographies, presentations, and review papers based on our motive and file the research.
- We can our scope to help others and publish us a skilled person in this topic.
The objective of this review is to crucially recognize and integrate the real-time content in the area. Though it is a time-consuming work, it will be useful for someone aims to make research and latest works in DL.
Deep Learning project face recognition with python OpenCV
Designing a face remembering system using Python and OpenCV is an amazing work that introduces us into the world of computer vision and DL. The following are the step-by-step guide to construct a simple face recognition system:
- Install Necessary Libraries
Make sure that we have the required libraries installed:
pip install opencv-python opencv-python-headless
- Capture Faces
We require a dataset for training. We utilize the pre-defined dataset and capture our own using OpenCV.
cam = cv2.VideoCapture(0)
detector = cv2.CascadeClassifier(cv2.data.haarcascades + ‘haarcascade_frontalface_default.xml’)
id = input(‘Enter user ID: ‘)
sampleNum = 0
while True:
ret, img = cam.read()
gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
faces = detector.detectMultiScale(gray, 1.3, 5)
for (x,y,w,h) in faces:
sampleNum += 1
cv2.imwrite(f”faces/User.{id}.{sampleNum}.jpg”, gray[y:y+h,x:x+w])
cv2.rectangle(img, (x,y), (x+w, y+h), (255,0,0), 2)
cv2.waitKey(100)
cv2.imshow(‘Capture’, img)
cv2.waitKey(1)
if sampleNum > 20: # capture 20 images
break
cam.release()
cv2.destroyAllWindows()
- Training the Recognizer
OpenCV has a built-in face recognizer. For this example, we’ll use the LBPH (Local Binary Pattern Histogram) face recognizer.
import numpy as np
from PIL import Image
path = ‘faces’
recognizer = cv2.face.LBPHFaceRecognizer_create()
def getImagesAndLabels(path):
imagePaths = [os.path.join(path,f) for f in os.listdir(path)]
faceSamples=[]
ids = []
for imagePath in imagePaths:
PIL_img = Image.open(imagePath).convert(‘L’)
img_numpy = np.array(PIL_img,’uint8′)
id = int(os.path.split(imagePath)[-1].split(“.”)[1])
faces = detector.detectMultiScale(img_numpy)
for (x,y,w,h) in faces:
faceSamples.append(img_numpy[y:y+h,x:x+w])
ids.append(id)
return faceSamples, np.array(ids)
faces,ids = getImagesAndLabels(path)
recognizer.train(faces, ids)
recognizer.save(‘trainer/trainer.yml’)
- Recognizing Faces
recognizer.read(‘trainer/trainer.yml’)
cascadePath = cv2.data.haarcascades + “haarcascade_frontalface_default.xml”
faceCascade = cv2.CascadeClassifier(cascadePath)
font = cv2.FONT_HERSHEY_SIMPLEX
minW = 0.1*cam.get(3)
minH = 0.1*cam.get(4)
faces = faceCascade.detectMultiScale(
gray,
scaleFactor=1.2,
minNeighbors=5,
minSize=(int(minW), int(minH)),
id, confidence = recognizer.predict(gray[y:y+h,x:x+w])
if (confidence < 100):
confidence = f” {round(100 – confidence)}%”
else:
id = “unknown”
cv2.putText(img, str(id), (x+5,y-5), font, 1, (255,255,255), 2)
cv2.putText(img, str(confidence), (x+5,y+h-5), font, 1, (255,255,0), 1)
cv2.imshow(‘Face Recognition’,img)
if cv2.waitKey(1) & 0xFF == ord(‘q’):
We have proper directories (faces and trainer) to design. It will be a basic face recognition system and can strengthen with DL models for better accuracy and robustness against various states in real-time. To achieve better accuracy in real-time conditions, we discover latest DL based techniques like FaceNet or pre-trained models from DL frameworks.
Deep learning MS Thesis topics
Have a conversation with our faculty members to get the best topics that matches with your interest. Some of the unique topic ideas are shared below …. contact us for more support.

- Modulation Recognition based on Incremental Deep Learning
- Fast Channel Analysis and Design Approach using Deep Learning Algorithm for 112Gbs HSI Signal Routing Optimization
- Deep Learning of Process Data with Supervised Variational Auto-encoder for Soft Sensor
- Methodological Principles for Deep Learning in Software Engineering
- Recent Trends in Deep Learning for Natural Language Processing and Scope for Asian Languages
- Adding Context to Source Code Representations for Deep Learning
- Weekly Power Generation Forecasting using Deep Learning Techniques: Case Study of a 1.5 MWp Floating PV Power Plant
- A Study of Deep Learning Approaches and Loss Functions for Abundance Fractions Estimation
- A Trustless Federated Framework for Decentralized and Confidential Deep Learning
- Research on Financial Data Analysis Based on Applied Deep Learning in Quantitative Trading
- A Deep Learning model for day-ahead load forecasting taking advantage of expert knowledge
- Locational marginal price forecasting using Transformer-based deep learning network
- H-Stegonet: A Hybrid Deep Learning Framework for Robust Steganalysis
- Comparison of Deep Learning Approaches for Sentiment Classification
- An Unmanned Network Intrusion Detection Model Based on Deep Reinforcement Learning
- Indoor Object Localization and Tracking Using Deep Learning over Received Signal Strength
- Analysis of Deep Learning 3-D Imaging Methods Based on UAV SAR
- Research and improvement of deep learning tool chain for electric power applications
- Hybrid Intrusion Detector using Deep Learning Technique
- Non-Trusted user Classification-Comparative Analysis of Machine and Deep Learning Approaches
Why Work With Us ?
Senior research member, research experience, journal member, book publisher, research ethics, business ethics, valid references, explanations, paper publication, 9 big reasons to select us.
Our Editor-in-Chief has Website Ownership who control and deliver all aspects of PhD Direction to scholars and students and also keep the look to fully manage all our clients.
Our world-class certified experts have 18+years of experience in Research & Development programs (Industrial Research) who absolutely immersed as many scholars as possible in developing strong PhD research projects.
We associated with 200+reputed SCI and SCOPUS indexed journals (SJR ranking) for getting research work to be published in standard journals (Your first-choice journal).
PhDdirection.com is world’s largest book publishing platform that predominantly work subject-wise categories for scholars/students to assist their books writing and takes out into the University Library.
Our researchers provide required research ethics such as Confidentiality & Privacy, Novelty (valuable research), Plagiarism-Free, and Timely Delivery. Our customers have freedom to examine their current specific research activities.
Our organization take into consideration of customer satisfaction, online, offline support and professional works deliver since these are the actual inspiring business factors.
Solid works delivering by young qualified global research team. "References" is the key to evaluating works easier because we carefully assess scholars findings.
Detailed Videos, Readme files, Screenshots are provided for all research projects. We provide Teamviewer support and other online channels for project explanation.
Worthy journal publication is our main thing like IEEE, ACM, Springer, IET, Elsevier, etc. We substantially reduces scholars burden in publication side. We carry scholars from initial submission to final acceptance.
Related Pages
Our benefits, throughout reference, confidential agreement, research no way resale, plagiarism-free, publication guarantee, customize support, fair revisions, business professionalism, domains & tools, we generally use, wireless communication (4g lte, and 5g), ad hoc networks (vanet, manet, etc.), wireless sensor networks, software defined networks, network security, internet of things (mqtt, coap), internet of vehicles, cloud computing, fog computing, edge computing, mobile computing, mobile cloud computing, ubiquitous computing, digital image processing, medical image processing, pattern analysis and machine intelligence, geoscience and remote sensing, big data analytics, data mining, power electronics, web of things, digital forensics, natural language processing, automation systems, artificial intelligence, mininet 2.1.0, matlab (r2018b/r2019a), matlab and simulink, apache hadoop, apache spark mlib, apache mahout, apache flink, apache storm, apache cassandra, pig and hive, rapid miner, support 24/7, call us @ any time, +91 9444829042, [email protected].
Questions ?
Click here to chat with us
- Warning : Invalid argument supplied for foreach() in /home/customer/www/opendatascience.com/public_html/wp-includes/nav-menu.php on line 95 Warning : array_merge(): Expected parameter 2 to be an array, null given in /home/customer/www/opendatascience.com/public_html/wp-includes/nav-menu.php on line 102
- ODSC EUROPE
- AI+ Training
- Speak at ODSC

- Data Analytics
- Data Engineering
- Data Visualization
- Deep Learning
- Generative AI
- Machine Learning
- NLP and LLMs
- Business & Use Cases
- Career Advice
- Write for us
- ODSC Community Slack Channel
- Upcoming Webinars

Best Deep Learning Research of 2021 So Far
Deep Learning Modeling Research posted by Daniel Gutierrez, ODSC August 2, 2021 Daniel Gutierrez, ODSC
The discipline of AI most often mentioned these days is deep learning (DL) along with its many incarnations implemented with deep neural networks. DL also is a rapidly accelerating area of research with papers being published at a fast clip by research teams from around the globe.
I enjoy keeping a pulse on deep learning research and so far in 2021 research innovations have propagated at a quick pace. Some of the top topical areas for deep learning research are: causality, explainability/interpretability, transformers, NLP, GPT, language models, GANs, deep learning for tabular data, and many others.
In this article, we’ll take a brief tour of my top picks for deep learning research (in no particular order) of papers that I found to be particularly compelling. I’m pretty attached to this leading-edge research. I’m known to carry a thick folder of recent research papers around in my backpack and consume all the great developments when I have a spare moment. Enjoy!
Check out my previous lists: Best Machine Learning Research of 2021 So Far , Best of Deep Reinforcement Learning Research of 2019 , Most Influential NLP Research of 2019 , and Most Influential Deep Learning Research of 2019 .
Cause and Effect: Concept-based Explanation of Neural Networks
In many scenarios, human decisions are explained based on some high-level concepts. This paper takes a step in the interpretability of neural networks by examining their internal representation or neuron’s activations against concepts. A concept is characterized by a set of samples that have specific features in common. A framework is proposed to check the existence of a causal relationship between a concept (or its negation) and task classes. While the previous methods focus on the importance of a concept to a task class, the paper goes further and introduces four measures to quantitatively determine the order of causality. Through experiments, the effectiveness of the proposed method is demonstrated in explaining the relationship between a concept and the predictive behavior of a neural network.
Pretrained Language Models for Text Generation: A Survey
Text generation has become one of the most important yet challenging tasks in natural language processing (NLP). The resurgence of deep learning has greatly advanced this field by neural generation models, especially the paradigm of pretrained language models (PLMs). This paper presents an overview of the major advances achieved in the topic of PLMs for text generation. As the preliminaries, the paper presents the general task definition and briefly describes the mainstream architectures of PLMs for text generation. As the core content, the deep learning research paper discusses how to adapt existing PLMs to model different input data and satisfy special properties in the generated text.
A Short Survey of Pre-trained Language Models for Conversational AI-A NewAge in NLP
Building a dialogue system that can communicate naturally with humans is a challenging yet interesting problem of agent-based computing. The rapid growth in this area is usually hindered by the long-standing problem of data scarcity as these systems are expected to learn syntax, grammar, decision making, and reasoning from insufficient amounts of task-specific data sets. The recently introduced pre-trained language models have the potential to address the issue of data scarcity and bring considerable advantages by generating contextualized word embeddings. These models are considered counterparts of ImageNet in NLP and have demonstrated the ability to capture different facets of language such as hierarchical relations, long-term dependency, and sentiment. This short survey paper discusses the recent progress made in the field of pre-trained language models.
TrustyAI Explainability Toolkit
AI is becoming increasingly more popular and can be found in workplaces and homes around the world. However, how do we ensure trust in these systems? Regulation changes such as the GDPR mean that users have a right to understand how their data has been processed as well as saved. Therefore if, for example, you are denied a loan you have the right to ask why. This can be hard if the method for working this out uses “black box” machine learning techniques such as neural networks. TrustyAI is a new initiative which looks into explainable artificial intelligence (XAI) solutions to address trustworthiness in ML as well as decision services landscapes. This deep learning research paper looks at how TrustyAI can support trust in decision services and predictive models. The paper investigates techniques such as LIME, SHAP and counterfactuals, benchmarking both LIME and counterfactual techniques against existing implementations.
Generative Adversarial Network: Some Analytical Perspectives
Ever since its debut, generative adversarial networks (GANs) have attracted tremendous amount of attention. Over the past years, different variations of GANs models have been developed and tailored to different applications in practice. Meanwhile, some issues regarding the performance and training of GANs have been noticed and investigated from various theoretical perspectives. This paper starts from an introduction of GANs from an analytical perspective, then moves onto the training of GANs via SDE approximations and finally discusses some applications of GANs in computing high dimensional MFGs as well as tackling mathematical finance problems.
PyTorch Tabular: A Framework for Deep Learning with Tabular Data
In spite of showing unreasonable effectiveness in modalities like Text and Image, deep learning has always lagged gradient boosting in tabular data – both in popularity and performance. But recently there have been newer models created specifically for tabular data, which is pushing the performance bar. But popularity is still a challenge because there is no easy, ready-to-use library like scikit-learn for deep learning. PyTorch Tabular is a new deep learning library which makes working with deep learning and tabular data easy and fast. It is a library built on top of PyTorch and PyTorch Lightning and works on Pandas dataframes directly. Many SOTA models like NODE and TabNet are already integrated and implemented in the library with a unified API. PyTorch Tabular is designed to be easily extensible for researchers, simple for practitioners, and robust in industrial deployments.
A Survey of Quantization Methods for Efficient Neural Network Inference
As soon as abstract mathematical computations were adapted to computation on digital computers, the problem of efficient representation, manipulation, and communication of the numerical values in those computations arose. Strongly related to the problem of numerical representation is the problem of quantization : in what manner should a set of continuous real-valued numbers be distributed over a fixed discrete set of numbers to minimize the number of bits required and also to maximize the accuracy of the attendant computations? This perennial problem of quantization is particularly relevant whenever memory and/or computational resources are severely restricted, and it has come to the forefront in recent years due to the remarkable performance of Neural Network models in computer vision, natural language processing, and related areas. Moving from floating-point representations to low-precision fixed integer values represented in four bits or less holds the potential to reduce the memory footprint and latency by a factor of 16x; and, in fact, reductions of 4x to 8x are often realized in practice in these applications. Thus, it is not surprising that quantization has emerged recently as an important and very active sub-area of research in the efficient implementation of computations associated with Neural Networks. This paper surveys approaches to the problem of quantizing the numerical values in deep Neural Network computations, covering the advantages/disadvantages of current methods.
How to decay your learning rate
Complex learning rate schedules have become an integral part of deep learning. This research finds empirically that common fine-tuned schedules decay the learning rate after the weight norm bounces. This leads to the proposal of ABEL : an automatic scheduler which decays the learning rate by keeping track of the weight norm. ABEL’s performance matches that of tuned schedules and is more robust with respect to its parameters. Through extensive experiments in vision, NLP, and RL, it is shown that if the weight norm does not bounce, it is possible to simplify schedules even further with no loss in performance. In such cases, a complex schedule has similar performance to a constant learning rate with a decay at the end of training.
GPT Understands, Too
While GPTs with traditional fine-tuning fail to achieve strong results on natural language understanding (NLU), this paper shows that GPTs can be better than or comparable to similar-sized BERTs on NLU tasks with a novel method P-tuning — which employs trainable continuous prompt embeddings. On the knowledge probing (LAMA) benchmark, the best GPT recovers 64% (P@1) of world knowledge without any additional text provided during test time, which substantially improves the previous best by 20+ percentage points. On the SuperGlue benchmark, GPTs achieve comparable and sometimes better performance to similar-sized BERTs in supervised learning. Importantly, it is found that P-tuning also improves BERTs’ performance in both few-shot and supervised settings while largely reducing the need for prompt engineering. Consequently, P-tuning outperforms the state-of-the-art approaches on the few-shot SuperGlue benchmark.
Understanding Robustness of Transformers for Image Classification
Deep Convolutional Neural Networks (CNNs) have long been the architecture of choice for computer vision tasks. Recently, Transformer-based architectures like Vision Transformer (ViT) have matched or even surpassed ResNets for image classification. However, details of the Transformer architecture — such as the use of non-overlapping patches — lead one to wonder whether these networks are as robust. This paper performs an extensive study of a variety of different measures of robustness of ViT models and compare the findings to ResNet baselines. Investigated is robustness to input perturbations as well as robustness to model perturbations. The paper finds that when pre-trained with a sufficient amount of data, ViT models are at least as robust as the ResNet counterparts on a broad range of perturbations. Also found is that Transformers are robust to the removal of almost any single layer, and that while activations from later layers are highly correlated with each other, they nevertheless play an important role in classification.
Improving DeepFake Detection Using Dynamic Face Augmentation
The creation of altered and manipulated faces has become more common due to the improvement of DeepFake generation methods. Simultaneously, we have seen the development of detection models for differentiating between a manipulated and original face from image or video content. We have observed that most publicly available DeepFake detection datasets have limited variations, where a single face is used in many videos, resulting in an oversampled training dataset. Due to this, deep neural networks tend to overfit to the facial features instead of learning to detect manipulation features of DeepFake content. As a result, most detection architectures perform poorly when tested on unseen data. This paper provides a quantitative analysis to investigate this problem and present a solution to prevent model overfitting due to the high volume of samples generated from a small number of actors.
An Evaluation of Edge TPU Accelerators for Convolutional Neural Networks
Edge TPUs are a domain of accelerators for low-power, edge devices and are widely used in various Google products such as Coral and Pixel devices. This paper first discusses the major microarchitectural details of Edge TPUs. This is followed by an extensive evaluation of three classes of Edge TPUs, covering different computing ecosystems that are either currently deployed in Google products or are the product pipeline. Building upon this extensive study, the paper discusses critical and interpretable microarchitectural insights about the studied classes of Edge TPUs. Mainly discussed is how Edge TPU accelerators perform across CNNs with different structures. Finally, the paper presents ongoing efforts in developing high-accuracy learned machine learning models to estimate the major performance metrics of accelerators such as latency and energy consumption. These learned models enable significantly faster (in the order of milliseconds) evaluations of accelerators as an alternative to time-consuming cycle-accurate simulators and establish an exciting opportunity for rapid hard-ware/software co-design.
Attention Models for Point Clouds in Deep Learning: A Survey
Recently, the advancement of 3D point clouds in deep learning has attracted intensive research in different application domains such as computer vision and robotic tasks. However, creating feature representation of robust, discriminative from unordered and irregular point clouds is challenging. The goal of this paper is to provide a comprehensive overview of the point clouds feature representation which uses attention models. More than 75+ key contributions in the recent three years are summarized in this survey, including the 3D objective detection, 3D semantic segmentation, 3D pose estimation, point clouds completion etc. Also provided are: a detailed characterization of (i) the role of attention mechanisms, (ii) the usability of attention models into different tasks, and (iii) the development trend of key technology.
Constrained Optimization for Training Deep Neural Networks Under Class Imbalance
Deep neural networks (DNNs) are notorious for making more mistakes for the classes that have substantially fewer samples than the others during training. Such class imbalance is ubiquitous in clinical applications and very crucial to handle because the classes with fewer samples most often correspond to critical cases (e.g., cancer) where misclassifications can have severe consequences. Not to miss such cases, binary classifiers need to be operated at high True Positive Rates (TPR) by setting a higher threshold but this comes at the cost of very high False Positive Rates (FPR) for problems with class imbalance. Existing methods for learning under class imbalance most often do not take this into account. This paper argues that prediction accuracy should be improved by emphasizing reducing FPRs at high TPRs for problems where misclassification of the positive samples are associated with higher cost. To this end, it’s posed the training of a DNN for binary classification as a constrained optimization problem and introduce a novel constraint that can be used with existing loss functions to enforce maximal area under the ROC curve (AUC). The resulting constrained optimization problem is solved using an Augmented Lagrangian method (ALM), where the constraint emphasizes reduction of FPR at high TPR. Results demonstrate that the proposed method almost always improves the loss functions it is used with by attaining lower FPR at high TPR and higher or equal AUC.
Deep Convolutional Neural Networks with Unitary Weights
While normalizations aim to fix the exploding and vanishing gradient problem in deep neural networks, they have drawbacks in speed or accuracy because of their dependency on the data set statistics. This paper is a comprehensive study of a novel method based on unitary synaptic weights derived from Lie Group to construct intrinsically stable neural systems. Here it’s shown that unitary convolutional neural networks deliver up to 32% faster inference speeds while maintaining competitive prediction accuracy. Unlike prior arts restricted to square synaptic weights, the paper expands the unitary networks to weights of any size and dimension.
TransGAN: Two Pure Transformers Can Make One Strong GAN, and That Can Scale Up
The recent explosive interest with transformers has suggested their potential to become powerful “universal” models for computer vision tasks, such as classification, detection, and segmentation. An important question is how much further transformers can go – are they ready to take some more notoriously difficult vision tasks, e.g., generative adversarial networks (GANs)? Driven by that curiosity, this paper conducts the first pilot study in building a GAN completely free of convolutions, using only pure transformer-based architectures. The proposed vanilla GAN architecture, dubbed TransGAN , consists of a memory-friendly transformer-based generator that progressively increases feature resolution while decreasing embedding dimension, and a patch-level discriminator that is also transformer-based. TransGAN is seen to notably benefit from data augmentations (more than standard GANs), a multi-task co-training strategy for the generator, and a locally initialized self-attention that emphasizes the neighborhood smoothness of natural images. Equipped with those findings, TransGAN can effectively scale up with bigger models and high-resolution image datasets. Specifically, the architecture achieves highly competitive performance compared to current state-of-the-art GANs based on convolutional backbones. The GitHub repo associated with this paper can be found HERE .

Deep Learning for Scene Classification: A Survey
Scene classification , aiming at classifying a scene image to one of the predefined scene categories by comprehending the entire image, is a longstanding, fundamental and challenging problem in computer vision. The rise of large-scale datasets, which constitute a dense sampling of diverse real-world scenes, and the renaissance of deep learning techniques, which learn powerful feature representations directly from big raw data, have been bringing remarkable progress in the field of scene representation and classification. To help researchers master needed advances in this field, the goal of this paper is to provide a comprehensive survey of recent achievements in scene classification using deep learning. More than 260 major publications are included in this survey covering different aspects of scene classification, including challenges, benchmark datasets, taxonomy, and quantitative performance comparisons of the reviewed methods. In retrospect of what has been achieved so far, this paper is concluded with a list of promising research opportunities.
Introducing and assessing the explainable AI (XAI) method: SIDU
Explainable Artificial Intelligence (XAI) has in recent years become a well-suited framework to generate human-understandable explanations of black box models. This paper presents a novel XAI visual explanation algorithm denoted SIDU that can effectively localize entire object regions responsible for prediction. The paper analyzes its robustness and effectiveness through various computational and human subject experiments. In particular, the SIDU algorithm is assessed using three different types of evaluations (Application, Human and Functionally-Grounded) to demonstrate its superior performance. The robustness of SIDU is further studied in presence of adversarial attack on black box models to better understand its performance.
Evolving Reinforcement Learning Algorithms
This paper proposes a method for meta-learning reinforcement learning algorithms by searching over the space of computational graphs which compute the loss function for a value-based model-free RL agent to optimize. The learned algorithms are domain-agnostic and can generalize to new environments not seen during training. The method can both learn from scratch and bootstrap off known existing algorithms, like DQN, enabling interpretable modifications which improve performance. Learning from scratch on simple classical control and gridworld tasks, the method rediscovers the temporal-difference (TD) algorithm. Bootstrapped from DQN, two learned algorithms are highlighted which obtain good generalization performance over other classical control tasks, gridworld type tasks, and Atari games. The analysis of the learned algorithm behavior shows resemblance to recently proposed RL algorithms that address overestimation in value-based methods.
RepVGG: Making VGG-style ConvNets Great Again
VGG-style ConvNets, although now considered a classic architecture, were attractive due to their simplicity. In contrast, ResNets have become popular due to their high accuracy but are more difficult to customize and display undesired inference drawbacks. To address these issues, Ding et al. propose RepVGG – the return of the VGG!
RepVGG is an efficient and simple architecture using plain VGG-style ConvNets. It decouples the inference-time and training-time architecture through a structural re-parameterization technique. The researchers report favorable speed-accuracy tradeoff compared to state-of-the-art models, such as EfficientNet and RegNet. RepVGG achieves 80% top-1 accuracy on ImageNet and is benchmarked as being 83% faster than ResNet-50. This research is part of a broader effort to build more efficient models using simpler architectures and operations. The GitHub repo associated with this paper can be found HERE .
Switch Transformers: Scaling to Trillion Parameter Models with Simple and Efficient Sparsity
In deep learning, models typically reuse the same parameters for all inputs. Mixture of Experts (MoE) defies this and instead selects different parameters for each incoming example. The result is a sparsely-activated model — with outrageous numbers of parameters — but a constant computational cost. However, despite several notable successes of MoE, widespread adoption has been hindered by complexity, communication costs and training instability — this paper addresses these with the Switch Transformer . The Google Brain researchers simplify the MoE routing algorithm and design intuitive improved models with reduced communication and computational costs. The proposed training techniques help wrangle the instabilities and it is shown that large sparse models may be trained, for the first time, with lower precision (bfloat16) formats. They design models based off T5-Base and T5-Large to obtain up to 7x increases in pre-training speed with the same computational resources. These improvements extend into multilingual settings to measure gains over the mT5-Base version across all 101 languages. Finally, the paper advances the current scale of language models by pre-training up to trillion parameter models on the “Colossal Clean Crawled Corpus” and achieve a 4x speedup over the T5-XXL model. The GitHub repo associated with this paper can be found HERE .
How to Learn More about Deep Learning Research
At our upcoming event this November 16th-18th in San Francisco, ODSC West 2021 will feature a plethora of talks, workshops, and training sessions on deep learning and deep learning research. You can register now for 60% off all ticket types before the discount drops to 40% in a few weeks. Some highlighted sessions on deep learning include:
Sessions on Deep Learning and Deep Learning Research:
- GANs: Theory and Practice, Image Synthesis With GANs Using TensorFlow: Ajay Baranwal | Center Director | Center for Deep Learning in Electronic Manufacturing, Inc
- Machine Learning With Graphs: Going Beyond Tabular Data: Dr. Clair J. Sullivan | Data Science Advocate | Neo4j
- Deep Dive into Reinforcement Learning with PPO using TF-Agents & TensorFlow 2.0: Oliver Zeigermann | Software Developer | embarc Software Consulting GmbH
- Get Started with Time-Series Forecasting using the Google Cloud AI Platform: Karl Weinmeister | Developer Relations Engineering Manager | Google
Sessions on Machine Learning:
- Towards More Energy-Efficient Neural Networks? Use Your Brain!: Olaf de Leeuw | Data Scientist | Dataworkz
- Practical MLOps: Automation Journey: Evgenii Vinogradov, PhD | Head of DHW Development | YooMoney
- Applications of Modern Survival Modeling with Python: Brian Kent, PhD | Data Scientist | Founder The Crosstab Kite
- Using Change Detection Algorithms for Detecting Anomalous Behavior in Large Systems: Veena Mendiratta, PhD | Adjunct Faculty, Network Reliability and Analytics Researcher | Northwestern University
Sessions on MLOps:
- Tuning Hyperparameters with Reproducible Experiments: Milecia McGregor | Senior Software Engineer | Iterative
- MLOps… From Model to Production: Filipa Peleja, PhD | Lead Data Scientist | Levi Strauss & Co
- Operationalization of Models Developed and Deployed in Heterogeneous Platforms: Sourav Mazumder | Data Scientist, Thought Leader, AI & ML Operationalization Leader | IBM
- Develop and Deploy a Machine Learning Pipeline in 45 Minutes with Ploomber: Eduardo Blancas | Data Scientist | Fidelity Investment

Daniel Gutierrez, ODSC
Daniel D. Gutierrez is a practicing data scientist who’s been working with data long before the field came in vogue. As a technology journalist, he enjoys keeping a pulse on this fast-paced industry. Daniel is also an educator having taught data science, machine learning and R classes at the university level. He has authored four computer industry books on database and data science technology, including his most recent title, “Machine Learning and Data Science: An Introduction to Statistical Learning Methods with R.” Daniel holds a BS in Mathematics and Computer Science from UCLA.

Tesla CEO Elon Musk Boosting Salaries in Bid to Fight Off OpenAI Poachers
AI and Data Science News posted by ODSC Team Apr 5, 2024 In a bid to fight off OpenAI poachers, Tesla CEO Elon Musk announced significant pay raises...

ODSC’s AI Weekly Recap: Week of April 5th
AI and Data Science News posted by Jorge Arenas Apr 5, 2024 Open Data Science Blog Recap The White House Office of Management and Budget, or OMB has...

White House Pushes Fed Agencies to Hire AI Chiefs
AI and Data Science News posted by ODSC Team Apr 4, 2024 The White House Office of Management and Budget, or OMB has introduced guidelines mandating all U.S....

deep learning Recently Published Documents
Total documents.
- Latest Documents
- Most Cited Documents
- Contributed Authors
- Related Sources
- Related Keywords
Synergic Deep Learning for Smart Health Diagnosis of COVID-19 for Connected Living and Smart Cities
COVID-19 pandemic has led to a significant loss of global deaths, economical status, and so on. To prevent and control COVID-19, a range of smart, complex, spatially heterogeneous, control solutions, and strategies have been conducted. Earlier classification of 2019 novel coronavirus disease (COVID-19) is needed to cure and control the disease. It results in a requirement of secondary diagnosis models, since no precise automated toolkits exist. The latest finding attained using radiological imaging techniques highlighted that the images hold noticeable details regarding the COVID-19 virus. The application of recent artificial intelligence (AI) and deep learning (DL) approaches integrated to radiological images finds useful to accurately detect the disease. This article introduces a new synergic deep learning (SDL)-based smart health diagnosis of COVID-19 using Chest X-Ray Images. The SDL makes use of dual deep convolutional neural networks (DCNNs) and involves a mutual learning process from one another. Particularly, the representation of images learned by both DCNNs is provided as the input of a synergic network, which has a fully connected structure and predicts whether the pair of input images come under the identical class. Besides, the proposed SDL model involves a fuzzy bilateral filtering (FBF) model to pre-process the input image. The integration of FBL and SDL resulted in the effective classification of COVID-19. To investigate the classifier outcome of the SDL model, a detailed set of simulations takes place and ensures the effective performance of the FBF-SDL model over the compared methods.
A deep learning approach for remote heart rate estimation
Weakly supervised spatial deep learning for earth image segmentation based on imperfect polyline labels.
In recent years, deep learning has achieved tremendous success in image segmentation for computer vision applications. The performance of these models heavily relies on the availability of large-scale high-quality training labels (e.g., PASCAL VOC 2012). Unfortunately, such large-scale high-quality training data are often unavailable in many real-world spatial or spatiotemporal problems in earth science and remote sensing (e.g., mapping the nationwide river streams for water resource management). Although extensive efforts have been made to reduce the reliance on labeled data (e.g., semi-supervised or unsupervised learning, few-shot learning), the complex nature of geographic data such as spatial heterogeneity still requires sufficient training labels when transferring a pre-trained model from one region to another. On the other hand, it is often much easier to collect lower-quality training labels with imperfect alignment with earth imagery pixels (e.g., through interpreting coarse imagery by non-expert volunteers). However, directly training a deep neural network on imperfect labels with geometric annotation errors could significantly impact model performance. Existing research that overcomes imperfect training labels either focuses on errors in label class semantics or characterizes label location errors at the pixel level. These methods do not fully incorporate the geometric properties of label location errors in the vector representation. To fill the gap, this article proposes a weakly supervised learning framework to simultaneously update deep learning model parameters and infer hidden true vector label locations. Specifically, we model label location errors in the vector representation to partially reserve geometric properties (e.g., spatial contiguity within line segments). Evaluations on real-world datasets in the National Hydrography Dataset (NHD) refinement application illustrate that the proposed framework outperforms baseline methods in classification accuracy.
Prediction of Failure Categories in Plastic Extrusion Process with Deep Learning
Hyperparameters tuning of faster r-cnn deep learning transfer for persistent object detection in radar images, a comparative study of automated legal text classification using random forests and deep learning, a semi-supervised deep learning approach for vessel trajectory classification based on ais data, an improved approach towards more robust deep learning models for chemical kinetics, power system transient security assessment based on deep learning considering partial observability, a multi-attention collaborative deep learning approach for blood pressure prediction.
We develop a deep learning model based on Long Short-term Memory (LSTM) to predict blood pressure based on a unique data set collected from physical examination centers capturing comprehensive multi-year physical examination and lab results. In the Multi-attention Collaborative Deep Learning model (MAC-LSTM) we developed for this type of data, we incorporate three types of attention to generate more explainable and accurate results. In addition, we leverage information from similar users to enhance the predictive power of the model due to the challenges with short examination history. Our model significantly reduces predictive errors compared to several state-of-the-art baseline models. Experimental results not only demonstrate our model’s superiority but also provide us with new insights about factors influencing blood pressure. Our data is collected in a natural setting instead of a setting designed specifically to study blood pressure, and the physical examination items used to predict blood pressure are common items included in regular physical examinations for all the users. Therefore, our blood pressure prediction results can be easily used in an alert system for patients and doctors to plan prevention or intervention. The same approach can be used to predict other health-related indexes such as BMI.
Export Citation Format
Share document.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Brief Bioinform
- v.19(6); 2018 Nov

Deep learning for healthcare: review, opportunities and challenges
Riccardo miotto.
1 Institute for Next Generation Healthcare, Department of Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai, New York, NY
2 Division of Health Informatics, Department of Healthcare Policy and Research at Weill Cornell Medicine at Cornell University, New York, NY
Shuang Wang
3 Department of Biomedical Informatics at the University of California San Diego, La Jolla, CA
Xiaoqian Jiang
Joel t dudley.
4 the Institute for Next Generation Healthcare and associate professor in the Department of Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai, New York, NY
Gaining knowledge and actionable insights from complex, high-dimensional and heterogeneous biomedical data remains a key challenge in transforming health care. Various types of data have been emerging in modern biomedical research, including electronic health records, imaging, -omics, sensor data and text, which are complex, heterogeneous, poorly annotated and generally unstructured. Traditional data mining and statistical learning approaches typically need to first perform feature engineering to obtain effective and more robust features from those data, and then build prediction or clustering models on top of them. There are lots of challenges on both steps in a scenario of complicated data and lacking of sufficient domain knowledge. The latest advances in deep learning technologies provide new effective paradigms to obtain end-to-end learning models from complex data. In this article, we review the recent literature on applying deep learning technologies to advance the health care domain. Based on the analyzed work, we suggest that deep learning approaches could be the vehicle for translating big biomedical data into improved human health. However, we also note limitations and needs for improved methods development and applications, especially in terms of ease-of-understanding for domain experts and citizen scientists. We discuss such challenges and suggest developing holistic and meaningful interpretable architectures to bridge deep learning models and human interpretability.
Introduction
Health care is coming to a new era where the abundant biomedical data are playing more and more important roles. In this context, for example, precision medicine attempts to ‘ensure that the right treatment is delivered to the right patient at the right time’ by taking into account several aspects of patient's data, including variability in molecular traits, environment, electronic health records (EHRs) and lifestyle [ 1–3 ].
The large availability of biomedical data brings tremendous opportunities and challenges to health care research. In particular, exploring the associations among all the different pieces of information in these data sets is a fundamental problem to develop reliable medical tools based on data-driven approaches and machine learning. To this aim, previous works tried to link multiple data sources to build joint knowledge bases that could be used for predictive analysis and discovery [ 4–6 ]. Although existing models demonstrate great promises (e.g. [ 7–11 ]), predictive tools based on machine learning techniques have not been widely applied in medicine [ 12 ]. In fact, there remain many challenges in making full use of the biomedical data, owing to their high-dimensionality, heterogeneity, temporal dependency, sparsity and irregularity [ 13–15 ]. These challenges are further complicated by various medical ontologies used to generalize the data (e.g. Systematized Nomenclature of Medicine-Clinical Terms (SNOMED-CT) [ 16 ], Unified Medical Language System (UMLS) [ 17 ], International Classification of Disease-9th version (ICD-9) [ 18 ]), which often contain conflicts and inconsistency [ 19 ]. Sometimes, the same clinical phenotype is also expressed in different ways across the data. As an example, in the EHRs, a patient diagnosed with ‘type 2 diabetes mellitus’ can be identified by laboratory values of hemoglobin A1C >7.0, presence of 250.00 ICD-9 code, ‘type 2 diabetes mellitus’ mentioned in the free-text clinical notes and so on. Consequently, it is nontrivial to harmonize all these medical concepts to build a higher-level semantic structure and understand their correlations [ 6 , 20 ].
A common approach in biomedical research is to have a domain expert to specify the phenotypes to use in an ad hoc manner. However, supervised definition of the feature space scales poorly and misses the opportunities to discover novel patterns. Alternatively, representation learning methods allow to automatically discover the representations needed for prediction from the raw data [ 21 , 22 ]. Deep learning methods are representation-learning algorithms with multiple levels of representation, obtained by composing simple but nonlinear modules that each transform the representation at one level (starting with the raw input) into a representation at a higher, slightly more abstract level [ 23 ]. Deep learning models demonstrated great performance and potential in computer vision, speech recognition and natural language processing tasks [ 24–27 ].
Given its demonstrated performance in different domains and the rapid progresses of methodological improvements, deep learning paradigms introduce exciting new opportunities for biomedical informatics. Efforts to apply deep learning methods to health care are already planned or underway. For example, Google DeepMind has announced plans to apply its expertise to health care [ 28 ] and Enlitic is using deep learning intelligence to spot health problems on X-rays and Computed Tomography (CT) scans [ 29 ].
However, deep learning approaches have not been extensively evaluated for a broad range of medical problems that could benefit from its capabilities. There are many aspects of deep learning that could be helpful in health care, such as its superior performance, end-to-end learning scheme with integrated feature learning, capability of handling complex and multi-modality data and so on. To accelerate these efforts, the deep learning research field as a whole must address several challenges relating to the characteristics of health care data (i.e. sparse, noisy, heterogeneous, time-dependent) as need for improved methods and tools that enable deep learning to interface with health care information workflows and clinical decision support.
In this article, we discuss recent and forthcoming applications of deep learning in medicine, highlighting the key aspects to significantly impact health care. We do not aim to provide a comprehensive background on technical details (see e.g. [ 21 , 30–32 ]) or general application of deep learning (see e.g. [ 23 ]). Instead, we focus on biomedical data only, in particular those originated from clinical imaging, EHRs, genomes and wearable devices. While additional sources of information, such as metabolome, antibodyome and other omics information are expected to be valuable for health monitoring, at this point deep learning has not been significantly used in these domains. Thus, in the following, we briefly introduce the general deep learning framework, we review some of its applications in the medical domain and we discuss the opportunities, challenges and applications related to these methods when used in the context of precision medicine and next-generation health care.
Deep learning framework
Machine learning is a general-purpose method of artificial intelligence that can learn relationships from the data without the need to define them a priori [ 33 ]. The major appeal is the ability to derive predictive models without a need for strong assumptions about the underlying mechanisms, which are usually unknown or insufficiently defined [ 34 ]. The typical machine learning workflow involves four steps: data harmonization, representation learning, model fitting and evaluation [ 35 ]. For decades, constructing a machine learning system required careful engineering and domain expertise to transform the raw data into a suitable internal representation from which the learning subsystem, often a classifier, could detect patterns in the data set. Conventional techniques are composed of a single, often linear, transformation of the input space and are limited in their ability to process natural data in their raw form [ 21 ].
Deep learning is different from traditional machine learning in how representations are learned from the raw data. In fact, deep learning allows computational models that are composed of multiple processing layers based on neural networks to learn representations of data with multiple levels of abstraction [ 23 ]. The major differences between deep learning and traditional artificial neural networks (ANNs) are the number of hidden layers, their connections and the capability to learn meaningful abstractions of the inputs. In fact, traditional ANNs are usually limited to three layers and are trained to obtain supervised representations that are optimized only for the specific task and are usually not generalizable [ 36 ]. Differently, every layer of a deep learning system produces a representation of the observed patterns based on the data it receives as inputs from the layer below, by optimizing a local unsupervised criterion [ 37 ]. The key aspect of deep learning is that these layers of features are not designed by human engineers, but they are learned from data using a general-purpose learning procedure. Figure 1 illustrates such differences at a high level: deep neural networks process the inputs in a layer-wise nonlinear manner to pre-train (initialize) the nodes in subsequent hidden layers to learn ‘deep structures’ and representations that are generalizable. These representations are then fed into a supervised layer to fine-tune the whole network using the backpropagation algorithm toward representations that are optimized for the specific end-to-end task.

Comparison between ANNs and deep architectures. While ANNs are usually composed by three layers and one transformation toward the final outputs, deep learning architectures are constituted by several layers of neural networks. Layer-wise unsupervised pre-training allows deep networks to be tuned efficiently and to extract deep structure from inputs to serve as higher-level features that are used to obtain better predictions.
The unsupervised pre-training breakthrough [ 23 , 38 ], new methods to prevent overfitting [ 39 ], the use of general-purpose graphic processing units to speedup computations and the development of high-level modules to easily build neural networks (e.g. Theano [ 40 ], Caffe [ 41 ], TensorFlow [ 42 ]) allowed deep models to establish as state-of-the-art solutions for several tasks. In fact, deep learning turned out to be good at discovering intricate structures in high-dimensional data and obtained remarkable performances for object detection in images [ 43 , 44 ], speech recognition [ 45 ] and natural language understanding [ 46 ] and translation [ 47 ]. Relevant clinical-ready successes have been obtained in health care as well (e.g. detection of diabetic retinopathy in retinal fundus photographs [ 48 ], classification of skin cancer [ 49 ], predicting of the sequence specificities of DNA- and RNA-binding proteins [ 50 ]), initiating the way toward a potential new generation of intelligent tools-based deep learning for real-world medical care.
Literature review
The use of deep learning for medicine is recent and not thoroughly explored. In the next sections, we will review some of the main recent literature (i.e. 32 papers) related to applications of deep models to clinical imaging, EHRs, genomics and wearable device data.
Table 1 summarizes all the papers mentioned in this literature review, in particular highlighting the type of networks and the medical data considered. To the best of our knowledge, there are no studies using deep learning to combine neither all these data sources, nor a part of them (e.g. only EHRs and clinical images, only EHRs and genomics) in a joint representation for medical analysis and prediction. A few preliminary studies evaluated the combined use of EHRs and genomics (e.g. see [ 9 , 80 ]), without applying deep learning though; for this reason, they were not considered relevant to this review. The deep architectures applied to the health care domain have been mostly based on convolutional neural networks (CNNs) [ 81 ], recurrent neural networks (RNNs) [ 82 ], Restricted Boltzmann Machines (RBMs) [ 83 ] and Autoencoders (AEs) [ 84 ]. Table 2 briefly reviews these models and provides the main ideas behind their structures.
Summary of the articles described in the literature review with highlighted the deep learning architecture applied and the medical domain considered
We report 32 different papers using deep learning on clinical images, EHRs, genomics and mobile data. As it can be seen, most of the papers apply CNNs and AEs, regardless the medical domain. To the best of our knowledge, no works in the literature jointly process these different types of data (e.g. all of them, only EHRs and clinical images, only EHRs and mobile data) using deep learning for medical intelligence and prediction.
RNN = recurrent neural network; CNN = convolutional neural network; RBM = restricted Boltzmann machine; AE = autoencoder; LSTM = long short-term memory; GRU = gated recurrent unit.
Review of the neural networks shaping the deep learning architectures applied to the health care domain in the literature
Clinical imaging
Following the success in computer vision, the first applications of deep learning to clinical data were on image processing, especially on the analysis of brain Magnetic Resonance Imaging (MRI) scans to predict Alzheimer disease and its variations [ 51 , 52 ]. In other medical domains, CNNs were used to infer a hierarchical representation of low-field knee MRI scans to automatically segment cartilage and predict the risk of osteoarthritis [ 53 ]. Despite using 2D images, this approach obtained better results than a state-of-the-art method using manually selected 3D multi-scale features. Deep learning was also applied to segment multiple sclerosis lesions in multi-channel 3D MRI [ 54 ] and for the differential diagnosis of benign and malignant breast nodules from ultrasound images [ 55 ]. More recently, Gulshan et al . [ 48 ] used CNNs to identify diabetic retinopathy in retinal fundus photographs, obtaining high sensitivity and specificity over about 10 000 test images with respect to certified ophthalmologist annotations. CNNs also obtained performances on par with 21 board-certified dermatologists on classifying biopsy-proven clinical images of different types of skin cancer (keratinocyte carcinomas versus benign seborrheic keratoses and malignant melanomas versus benign nevi) over a large data set of 130 000 images (1942 biopsy-labeled test images) [ 49 ].
Electronic health records
More recently deep learning has been applied to process aggregated EHRs, including both structured (e.g. diagnosis, medications, laboratory tests) and unstructured (e.g. free-text clinical notes) data. The greatest part of this literature processed the EHRs of a health care system with a deep architecture for a specific, usually supervised, predictive clinical task. In particular, a common approach is to show that deep learning obtains better results than conventional machine learning models with respect to certain metrics, such as Area Under the Receiver Operating Characteristic Curve, accuracy and F-score [ 91 ]. In this scenario, while most papers present end-to-end supervised networks, some works also propose unsupervised models to derive latent patient representations, which are then evaluated using shallow classifiers (e.g. random forests, logistic regression).
Several works applied deep learning to predict diseases from the patient clinical status. Liu et al . [ 56 ] used a four-layer CNN to predict congestive heart failure and chronic obstructive pulmonary disease and showed significant advantages over the baselines. RNNs with long short-term memory (LSTM) hidden units, pooling and word embedding were used in DeepCare [ 58 ], an end-to-end deep dynamic network that infers current illness states and predicts future medical outcomes. The authors also proposed to moderate the LSTM unit with a decay effect to handle irregular timed events (which are typical in longitudinal EHRs). Moreover, they incorporated medical interventions in the model to dynamically shape the predictions. DeepCare was evaluated for disease progression modeling, intervention recommendation and future risk prediction on diabetes and mental health patient cohorts. RNNs with gated recurrent unit (GRU) were used by Choi et al . [ 65 ] to develop Doctor AI, an end-to-end model that uses patient history to predict diagnoses and medications for subsequent encounters. The evaluation showed significantly higher recall than shallow baselines and good generalizability by adapting the resulting model from one institution to another without losing substantial accuracy. Differently, Miotto et al . [ 59 ] proposed to learn deep patient representations from the EHRs using a three-layer Stacked Denoising Autoencoder (SDA). They applied this novel representation on disease risk prediction using random forest as classifiers. The evaluation was performed on 76 214 patients comprising 78 diseases from diverse clinical domains and temporal windows (up to a 1 year). The results showed that the deep representation leads to significantly better predictions than using raw EHRs or conventional representation learning algorithms (e.g. Principal Component Analysis (PCA), k-means). Moreover, they also showed that results significantly improve when adding a logistic regression layer on top of the last AE to fine-tune the entire supervised network [ 60 ]. Similarly, Liang et al . [ 61 ] used RBMs to learn representations from EHRs that revealed novel concepts and demonstrated better prediction accuracy on a number of diseases.
Deep learning was also applied to model continuous time signals, such as laboratory results, toward the automatic identification of specific phenotypes. For example, Lipton et al . [ 57 ] used RNNs with LSTM to recognize patterns in multivariate time series of clinical measurements. Specifically, they trained a model to classify 128 diagnoses from 13 frequently but irregularly sampled clinical measurements from patients in pediatric intensive unit care. The results showed significant improvements with respect to several strong baselines, including multilayer perceptron trained on hand-engineered features. Che et al . [ 63 ] used SDAs regularized with a prior knowledge based on ICD-9s for detecting characteristic patterns of physiology in clinical time series. Lasko et al . [ 64 ] used a two-layer stacked AE (without regularization) to model longitudinal sequences of serum uric acid measurements to distinguish the uric-acid signatures of gout and acute leukemia. Razavian et al . [ 67 ] evaluated CNNs and RNNs with LSTM units to predict disease onset from laboratory test measures alone, showing better performances than logistic regression with hand-engineered, clinically relevant features.
Neural language deep models were also applied to EHRs, in particular to learn embedded representations of medical concepts, such as diseases, medications and laboratory tests, that could be used for analysis and prediction [ 92 ]. As an example, Tran et al . [ 62 ] used RBMs to learn abstractions of ICD-10 codes on a cohort of 7578 mental health patients to predict suicide risk. A deep architecture based on RNNs also obtained promising results in removing protected health information from clinical notes to leverage the automatic de-identification of free-text patient summaries [ 68 ].
The prediction of unplanned patient readmissions after discharge recently received attention as well. In this domain, Nguyen et al . [ 66 ] proposed Deepr, an end-to-end architecture based on CNNs, which detects and combines clinical motifs in the longitudinal patient EHRs to stratify medical risks. Deepr performed well in predicting readmission within 6 months and was able to detect meaningful and interpretable clinical patterns.
Deep learning in high-throughput biology is used to capture the internal structure of increasingly larger and high-dimensional data sets (e.g. DNA sequencing, RNA measurements). Deep models enable the discovery of high-level features, improving performances over traditional models, increasing interpretability and providing additional understanding about the structure of the biological data. Different works have been proposed in the literature. Here we review the general ideas and refer the reader to [ 93–96 ] for more comprehensive reviews.
The first applications of neural networks in genomics replaced conventional machine learning with deep architectures, without changing the input features. For example, Xiong et al . [ 97 ] used a fully connected feed-forward neural network to predict the splicing activity of individual exons. The model was trained using >1000 predefined features extracted from the candidate exon and adjacent introns. This method obtained higher prediction accuracy of splicing activity compared with simpler approaches, and was able to identify rare mutations implicated in splicing misregulation.
More recent works apply CNNs directly on the raw DNA sequence, without the need to define features a priori (e.g. [ 50 , 69 , 70 ]). CNNs use less parameters than a fully connected network by computing convolution on small regions of the input space and by sharing parameters between regions. This allowed training the models on larger sequence windows of DNAs, improving the detection of the relevant patterns. For example, Alipanahi et al . [ 50 ] proposed DeepBind, a deep architecture based on CNNs that predicts specificities of DNA- and RNA-binding proteins. In the reported experiment, DeepBind was able to recover known and novel sequence of motifs, quantify the effect of sequence alterations and identify functional single nucleotide variations (SNVs). Zhou and Troyanskaya [ 69 ] used CNNs to predict chromatin marks from DNA sequence. Similarly, Kelley et al . [ 70 ] developed Basset, an open-source framework to predict DNase I hypersensitivity across multiple cell types and to quantify the effect of SNVs on chromatin accessibility. CNNs were also used by Angermueller et al . [ 71 ] to predict DNA methylation states in single-cell bisulfite sequencing studies and, more recently, by Koh et al . [ 72 ] to denoise genome-wide chromatin immunoprecipitation followed by sequencing data to obtain a more accurate prevalence estimate for different chromatin marks.
While CNNs are the most widely used architectures to extract features from fixed-size DNA sequence windows, other deep architectures have been proposed as well. For example, sparse AEs were applied to classify cancer cases from gene expression profiles or to predict protein backbones [ 74 ]. Deep neural networks also enabled researchers to significantly improve the state-of-the-art drug discovery pipeline for genomic medicine [ 98 ].
Sensor-equipped smartphones and wearables are transforming a variety of mobile apps, including health monitoring [ 99 ]. As the difference between consumer health wearables and medical devices begins to soften, it is now possible for a single wearable device to monitor a range of medical risk factors. Potentially, these devices could give patients direct access to personal analytics that can contribute to their health, facilitate preventive care and aid in the management of ongoing illness [ 100 ]. Deep learning is considered to be a key element in analyzing this new type of data. However, only a few recent works used deep models within the health care sensing domain, mostly owing to hardware limitations. In fact, running an efficient and reliable deep architecture on a mobile device to process noisy and complex sensor data is still a challenging task that is likely to drain the device resources [ 101 ]. Several studies investigated solutions to overcome such hardware limitations. As an example, Lane and Georgiev [ 102 ] proposed a low-power deep neural network inference engine that exploited both Central Processing Unit (CPU) and Digital Signal Processor (DSP) of the mobile device, without leading to any major overburden of the hardware. They also proposed DeepX, a software accelerator capable of lowering the device resources required by deep learning that currently act as a severe bottleneck to mobile adoption. This architecture enabled large-scale deep learning to execute efficiently on mobile devices and significantly outperformed cloud-based off-loading solutions [ 103 ].
We did not find any relevant study applying deep learning on commercial wearable devices for health monitoring. However, a few works processed data from phones and medical monitor devices. In particular, relevant studies based on deep learning were done on Human Activity Recognition (HAR). While not directly exploring a medical application, many studies argue that the accurate predictions obtained by deep models on HAR can leverage clinical applications as well. In the health care domain, Hammerla et al . [ 75 ] evaluated CNNs and RNNs with LSTM to predict the freezing of gait in Parkinson disease (PD) patients. Freezing is a common motor complication in PD, where affected individuals struggle to initiate movements such as walking. Results based on accelerometer data from above the ankle, above the knee and on the trunk of 10 patients showed that RNNs obtained the best results, with a significantly large improvement over the other models, including CNNs. While the size of this data set was small, this study highlights the potential of deep learning in processing activity recognition measures for clinical use. Zhu et al . [ 76 ] obtained promising results in predicting Energy Expenditure (EE) from triaxial accelerometer and heart rate sensor data during ambulatory activities. EE is considered important in tracking personal activity and preventing chronic diseases such as obesity, diabetes and cardiovascular diseases. They used CNNs and significantly improved performances over regression and a shallow neural network.
In other clinical domains, deep learning, in particular CNNs and RBMs, improved over conventional machine learning in analyzing portable neurophysiological signals such as Electroencephalogram, Local Field Potentials and Photoplethysmography [ 77 , 78 ]. Differently, Sathyanarayana et al . [ 79 ] applied deep learning to predict poor or good sleep using actigraphy measurements of the physical activity of patients during awake time. In particular, by using a data set of 92 adolescents and one full week of monitored data, they showed that CNNs were able to obtain the highest specificity and sensitivity, with results 46% better than logistic regression.
Challenges and opportunities
Despite the promising results obtained using deep architectures, there remain several unsolved challenges facing the clinical application of deep learning to health care. In particular, we highlight the following key issues:
- Data volume : Deep learning refers to a set of highly intensive computational models. One typical example is fully connected multi-layer neural networks, where tons of network parameters need to be estimated properly. The basis to achieve this goal is the availability of huge amount of data. In fact, while there are no hard guidelines about the minimum number of training documents, a general rule of thumb is to have at least about 10× the number of samples as parameters in the network. This is also one of the reasons why deep learning is so successful in domains where huge amount of data can be easily collected (e.g. computer vision, speech, natural language). However, health care is a different domain; in fact, we only have approximately 7.5 billion people all over the world (as per September 2016), with a great part not having access to primary health care. Consequently, we cannot get as many patients as we want to train a comprehensive deep learning model. Moreover, understanding diseases and their variability is much more complicated than other tasks, such as image or speech recognition. Consequently, from a big data perspective, the amount of medical data that is needed to train an effective and robust deep learning model would be much more comparing with other media.
- Data quality : Unlike other domains where the data are clean and well-structured, health care data are highly heterogeneous, ambiguous, noisy and incomplete. Training a good deep learning model with such massive and variegate data sets is challenging and needs to consider several issues, such as data sparsity, redundancy and missing values.
- Temporality : The diseases are always progressing and changing over time in a nondeterministic way. However, many existing deep learning models, including those already proposed in the medical domain, assume static vector-based inputs, which cannot handle the time factor in a natural way. Designing deep learning approaches that can handle temporal health care data is an important aspect that will require the development of novel solutions.
- Domain complexity : Different from other application domains (e.g. image and speech analysis), the problems in biomedicine and health care are more complicated. The diseases are highly heterogeneous and for most of the diseases there is still no complete knowledge on their causes and how they progress. Moreover, the number of patients is usually limited in a practical clinical scenario and we cannot ask for as many patients as we want.
- Interpretability : Although deep learning models have been successful in quite a few application domains, they are often treated as black boxes. While this might not be a problem in other more deterministic domains such as image annotation (because the end user can objectively validate the tags assigned to the images), in health care, not only the quantitative algorithmic performance is important, but also the reason why the algorithms works is relevant. In fact, such model interpretability (i.e. providing which phenotypes are driving the predictions) is crucial for convincing the medical professionals about the actions recommended from the predictive system (e.g. prescription of a specific medication, potential high risk of developing a certain disease).
All these challenges introduce several opportunities and future research possibilities to improve the field. Therefore, with all of them in mind, we point out the following directions, which we believe would be promising for the future of deep learning in health care.
- Feature enrichment : Because of the limited amount of patients in the world, we should capture as many features as possible to characterize each patient and find novel methods to jointly process them. The data sources for generating those features need to include, but not to be limited to, EHRs, social media (e.g. there are prior research leveraging patient-reported information on social media for pharmacovigilance [ 104 , 105 ]), wearable devices, environments, surveys, online communities, genome profiles, omics data such as proteome and so on. The effective integration of such highly heterogeneous data and how to use them in a deep learning model would be an important and challenging research topic. In fact, to the best of our knowledge, the literature does not provide any study that attempts to combine different types of medical data sources using deep learning. A potential solution in this domain could exploit the hierarchical nature of deep learning and process separately every data source with the appropriate deep model, and stack the resulting representations in a joint model toward a holistic abstraction of the patient data (e.g. using layers of AEs or deep Bayesian networks).
- Federated inference : Each clinical institution possesses its own patient population. Building a deep learning model by leveraging the patients from different sites without leaking their sensitive information becomes a crucial problem in this setting. Consequently learning deep model in this federated setting in a secure way will be another important research topic, which will interface with other mathematical domains, such as cryptography (e.g. homomorphic encryption [ 106 ] and secure multiparty computation [ 107 ]).
- Model privacy : Privacy is an important concern in scaling up deep learning (e.g. through cloud computing services). In fact, a recent work by Tramèr et al . [ 108 ] has demonstrated the vulnerability of Machine Learning (ML)-as-a-service (i.e. ‘predictive analytics’) on a set of common models including deep neural networks. The attack abides all authentication or access-control mechanisms but infers parameters or training data through exposed Application Program Interface (APIs), which breaks the model and personal privacy. This issue is well known to the privacy community, and researchers have developed a principled framework called ‘differential privacy’ [ 109 , 110 ] to ensure the indistinguishability of individual samples in training data through their functional outputs [ 111 ]. However, naive approaches might render outputs useless or cannot provide sufficient protection [ 22 ], which makes the development of practically useful differential privacy solutions nontrivial. For example, Chaudhuri et al . [ 112 ] developed differential private methods to protect the parameters trained for the logistic regression model. Preserving the privacy of deep learning models is even more challenging, as there are more parameters to be safeguarded and several recent works have pushed the fronts in this area [ 113–115 ]. Yet, considering all the personal information likely to be processed by deep models in clinical applications, the deployment of intelligent tools for next-generation health care needs to consider these risks and attempt to implement a differential privacy standard.
- Incorporating expert knowledge : The existing expert knowledge for medical problems is invaluable for health care problems. Because of the limited amount of medical data and their various quality problems, incorporating the expert knowledge into the deep learning process to guide it toward the right direction is an important research topic. For example, online medical encyclopedia and PubMed abstracts should be mined to extract reliable content that can be included in the deep architecture to leverage the overall performances of the systems. Also semi-supervised learning, an effective scheme to learn from large amount of unlabeled samples with only a few labeled samples, would be of great potential because of its capability of leveraging both labeled (which encodes the knowledge) and unlabeled samples [ 105 ].
- Temporal modeling : Considering that the time factor is important in all kinds of health care-related problems, in particular in those involving EHRs and monitoring devices, training a time-sensitive deep learning model is critical for a better understanding of the patient condition and for providing timely clinical decision support. Thus, temporal deep learning is crucial for solving health care problems (as already shown in some of the early studies reported in the literature review). To this aim, we expect that RNNs as well as architectures coupled with memory (e.g. [ 86 ]) and attention mechanisms (e.g. [ 116 ]) will play a more significant role toward better clinical deep architectures.
- Interpretable modeling : Model performance and interpretability are equally important for health care problems. Clinicians are unlikely to adopt a system they cannot understand. Deep learning models are popular because of their superior performance. Yet, how to explain the results obtained by these models and how to make them more understandable is of key importance toward the development of trustable and reliable systems. In our opinion, research directions will include both algorithms to explain the deep models (i.e. what drives the hidden units of the networks to turn on/off along the process—see e.g. [ 117 ]) as well as methods to support the networks with existing tools that explain the predictions of data-driven systems (e.g. see [ 118 ]).
Applications
Deep learning methods are powerful tools that complement traditional machine learning and allow computers to learn from the data, so that they can come up with ways to create smarter applications. These approaches have already been used in a number of applications, especially for computer vision and natural language processing. All the results available in the literature illustrate the capabilities of deep learning for health care data analysis as well. In fact, processing medical data with multi-layer neural networks increased the predictive power for several specific applications in different clinical domains. Additionally, because of their hierarchical learning structure, deep architectures have the potential to integrate diverse data sets across heterogeneous data types and provide greater generalization given the focus on representation learning and not simply on classification accuracy.
Consequently, we believe that deep learning can open the way toward the next generation of predictive health care systems that can (i) scale to include many millions to billions of patient records and (ii) use a single, distributed patient representation to effectively support clinicians in their daily activities—rather than multiple systems working with different patient representations and data. Ideally, this representation would join all the different data sources, including EHRs, genomics, environment, wearables, social activities and so on, toward a holistic and comprehensive description of an individual status. In this scenario, the deep learning framework would be deployed into a health care platform (e.g. a hospital EHR system) and the models would be constantly updated to follow the changes in the patient population.
Such deep representations can then be used to leverage clinician activities in different domains and applications, such as disease risk prediction, personalized prescriptions, treatment recommendations, clinical trial recruitment as well as research and data analysis. As an example, Wang et al . recently won the Parkinson’s Progression Marker’s Initiative data challenge on subtyping Parkinson’s disease using a temporal deep learning approach [ 119 ]. In fact, because Parkinson’s disease is highly progressive, the traditional vector or matrix-based approach may not be optimal, as it is unable to accurately capture the disease progression patterns, as the entries in those vectors/matrices are typically aggregated over time. Consequently, the authors used the LSTM RNN model and identified three interesting subtypes for Parkinson’s disease, wherein each subtype demonstrates common disease progression trends. We believe that this work shows the great potential of deep learning models in real-world health care problems and how it could lead to more reliable and robust automatic systems in the near future.
Last, more broadly, deep learning can serve as a guiding principle to organize both hypothesis-driven research and exploratory investigation in clinical domains (e.g. clustering, visualization of patient cohorts, stratification of disease populations). For this potential to be realized, statistical and medical tasks must be integrated at all levels, including study design, experiment planning, model building and refinement and data interpretation.
- The fastest growing types of data in biomedical research, such as EHRs, imaging, -omics profiles and monitor data, are complex, heterogeneous, poorly annotated and generally unstructured.
- Early applications of deep learning to biomedical data showed effective opportunities to model, represent and learn from such complex and heterogeneous sources.
- State-of-the-art deep learning approaches need to be improved in terms of data integration, interpretability, security and temporal modeling to be effectively applied to the clinical domain.
- Deep learning can open the way toward the next generation of predictive health care systems, which can scale to include billions of patient records and rely on a single holistic patient representation to effectively support clinicians in their daily activities.
- Deep learning can serve as a guiding principle to organize both hypothesis-driven research and exploratory investigation in clinical domains based on different sources of data.
This study was supported by the following grants from the National Institute of Health: National Human Genome Research Institute (R00-HG008175) to S.W.; and National Library of Medicine (R21-LM012060); National Institute of Biomedical Imaging & Bioengineering (U01EB023685); (R01GM118609) to X.J.; National Institute of Diabetes and Digestive and Kidney Diseases (R01-DK098242-03); National Cancer Institute (U54-CA189201-02); and National Center for Advancing Translational Sciences (UL1TR000067) Clinical and Translational Science Awards to J.T.D. This study was also supported by the National Science Foundation: Information and Intelligent Systems (1650723) to F.W. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Biographies
Riccardo Miotto , PhD, is a senior data scientist in the Institute for Next Generation Healthcare, Department of Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai, New York, NY.
Fei Wang , PhD, is an assistant professor in the Division of Health Informatics, Department of Healthcare Policy and Research at Weill Cornell Medicine at Cornell University, New York, NY.
Shuang Wang , PhD, is an assistant professor in the Department of Biomedical Informatics at the University of California San Diego, La Jolla, CA.
Xiaoqian Jiang is an assistant professor in the Department of Biomedical Informatics at the University of California San Diego, La Jolla, CA.
Joel T. Dudley , PhD, is the executive director of the Institute for Next Generation Healthcare and associate professor in the Department of Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai, New York, NY.
Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, generate accurate citations for free.
- Knowledge Base
- Starting the research process
- How to Write a Research Proposal | Examples & Templates
How to Write a Research Proposal | Examples & Templates
Published on October 12, 2022 by Shona McCombes and Tegan George. Revised on November 21, 2023.
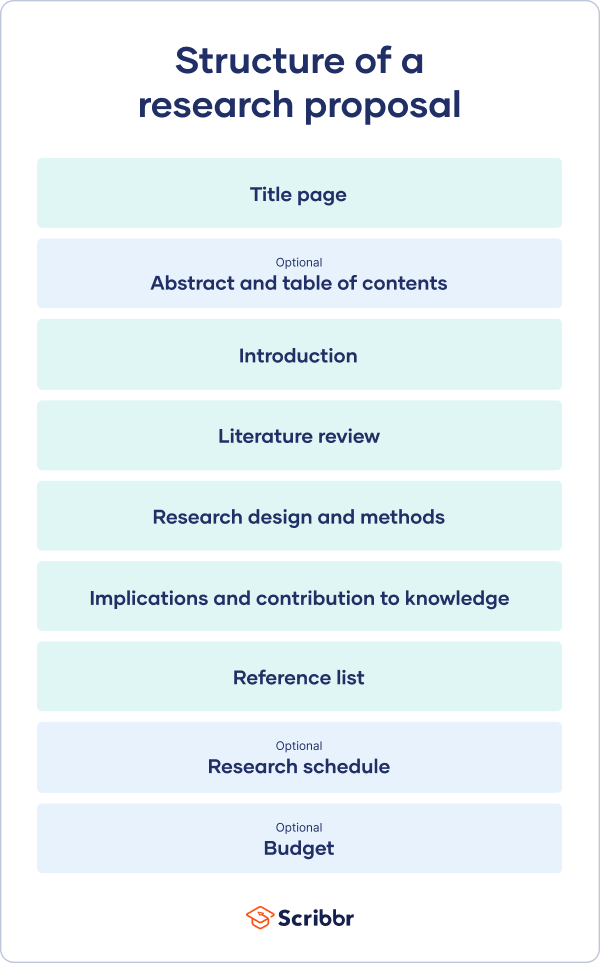
A research proposal describes what you will investigate, why it’s important, and how you will conduct your research.
The format of a research proposal varies between fields, but most proposals will contain at least these elements:
Introduction
Literature review.
- Research design
Reference list
While the sections may vary, the overall objective is always the same. A research proposal serves as a blueprint and guide for your research plan, helping you get organized and feel confident in the path forward you choose to take.
Table of contents
Research proposal purpose, research proposal examples, research design and methods, contribution to knowledge, research schedule, other interesting articles, frequently asked questions about research proposals.
Academics often have to write research proposals to get funding for their projects. As a student, you might have to write a research proposal as part of a grad school application , or prior to starting your thesis or dissertation .
In addition to helping you figure out what your research can look like, a proposal can also serve to demonstrate why your project is worth pursuing to a funder, educational institution, or supervisor.

Research proposal length
The length of a research proposal can vary quite a bit. A bachelor’s or master’s thesis proposal can be just a few pages, while proposals for PhD dissertations or research funding are usually much longer and more detailed. Your supervisor can help you determine the best length for your work.
One trick to get started is to think of your proposal’s structure as a shorter version of your thesis or dissertation , only without the results , conclusion and discussion sections.
Download our research proposal template
Receive feedback on language, structure, and formatting
Professional editors proofread and edit your paper by focusing on:
- Academic style
- Vague sentences
- Style consistency
See an example
Writing a research proposal can be quite challenging, but a good starting point could be to look at some examples. We’ve included a few for you below.
- Example research proposal #1: “A Conceptual Framework for Scheduling Constraint Management”
- Example research proposal #2: “Medical Students as Mediators of Change in Tobacco Use”
Like your dissertation or thesis, the proposal will usually have a title page that includes:
- The proposed title of your project
- Your supervisor’s name
- Your institution and department
The first part of your proposal is the initial pitch for your project. Make sure it succinctly explains what you want to do and why.
Your introduction should:
- Introduce your topic
- Give necessary background and context
- Outline your problem statement and research questions
To guide your introduction , include information about:
- Who could have an interest in the topic (e.g., scientists, policymakers)
- How much is already known about the topic
- What is missing from this current knowledge
- What new insights your research will contribute
- Why you believe this research is worth doing
Here's why students love Scribbr's proofreading services
Discover proofreading & editing
As you get started, it’s important to demonstrate that you’re familiar with the most important research on your topic. A strong literature review shows your reader that your project has a solid foundation in existing knowledge or theory. It also shows that you’re not simply repeating what other people have already done or said, but rather using existing research as a jumping-off point for your own.
In this section, share exactly how your project will contribute to ongoing conversations in the field by:
- Comparing and contrasting the main theories, methods, and debates
- Examining the strengths and weaknesses of different approaches
- Explaining how will you build on, challenge, or synthesize prior scholarship
Following the literature review, restate your main objectives . This brings the focus back to your own project. Next, your research design or methodology section will describe your overall approach, and the practical steps you will take to answer your research questions.
To finish your proposal on a strong note, explore the potential implications of your research for your field. Emphasize again what you aim to contribute and why it matters.
For example, your results might have implications for:
- Improving best practices
- Informing policymaking decisions
- Strengthening a theory or model
- Challenging popular or scientific beliefs
- Creating a basis for future research
Last but not least, your research proposal must include correct citations for every source you have used, compiled in a reference list . To create citations quickly and easily, you can use our free APA citation generator .
Some institutions or funders require a detailed timeline of the project, asking you to forecast what you will do at each stage and how long it may take. While not always required, be sure to check the requirements of your project.
Here’s an example schedule to help you get started. You can also download a template at the button below.
Download our research schedule template
If you are applying for research funding, chances are you will have to include a detailed budget. This shows your estimates of how much each part of your project will cost.
Make sure to check what type of costs the funding body will agree to cover. For each item, include:
- Cost : exactly how much money do you need?
- Justification : why is this cost necessary to complete the research?
- Source : how did you calculate the amount?
To determine your budget, think about:
- Travel costs : do you need to go somewhere to collect your data? How will you get there, and how much time will you need? What will you do there (e.g., interviews, archival research)?
- Materials : do you need access to any tools or technologies?
- Help : do you need to hire any research assistants for the project? What will they do, and how much will you pay them?
If you want to know more about the research process , methodology , research bias , or statistics , make sure to check out some of our other articles with explanations and examples.
Methodology
- Sampling methods
- Simple random sampling
- Stratified sampling
- Cluster sampling
- Likert scales
- Reproducibility
Statistics
- Null hypothesis
- Statistical power
- Probability distribution
- Effect size
- Poisson distribution
Research bias
- Optimism bias
- Cognitive bias
- Implicit bias
- Hawthorne effect
- Anchoring bias
- Explicit bias
Once you’ve decided on your research objectives , you need to explain them in your paper, at the end of your problem statement .
Keep your research objectives clear and concise, and use appropriate verbs to accurately convey the work that you will carry out for each one.
I will compare …
A research aim is a broad statement indicating the general purpose of your research project. It should appear in your introduction at the end of your problem statement , before your research objectives.
Research objectives are more specific than your research aim. They indicate the specific ways you’ll address the overarching aim.
A PhD, which is short for philosophiae doctor (doctor of philosophy in Latin), is the highest university degree that can be obtained. In a PhD, students spend 3–5 years writing a dissertation , which aims to make a significant, original contribution to current knowledge.
A PhD is intended to prepare students for a career as a researcher, whether that be in academia, the public sector, or the private sector.
A master’s is a 1- or 2-year graduate degree that can prepare you for a variety of careers.
All master’s involve graduate-level coursework. Some are research-intensive and intend to prepare students for further study in a PhD; these usually require their students to write a master’s thesis . Others focus on professional training for a specific career.
Critical thinking refers to the ability to evaluate information and to be aware of biases or assumptions, including your own.
Like information literacy , it involves evaluating arguments, identifying and solving problems in an objective and systematic way, and clearly communicating your ideas.
The best way to remember the difference between a research plan and a research proposal is that they have fundamentally different audiences. A research plan helps you, the researcher, organize your thoughts. On the other hand, a dissertation proposal or research proposal aims to convince others (e.g., a supervisor, a funding body, or a dissertation committee) that your research topic is relevant and worthy of being conducted.
Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the “Cite this Scribbr article” button to automatically add the citation to our free Citation Generator.
McCombes, S. & George, T. (2023, November 21). How to Write a Research Proposal | Examples & Templates. Scribbr. Retrieved April 3, 2024, from https://www.scribbr.com/research-process/research-proposal/
Is this article helpful?
Shona McCombes
Other students also liked, how to write a problem statement | guide & examples, writing strong research questions | criteria & examples, how to write a literature review | guide, examples, & templates, unlimited academic ai-proofreading.
✔ Document error-free in 5minutes ✔ Unlimited document corrections ✔ Specialized in correcting academic texts
Help | Advanced Search
Computer Science > Machine Learning
Title: deep generative models through the lens of the manifold hypothesis: a survey and new connections.
Abstract: In recent years there has been increased interest in understanding the interplay between deep generative models (DGMs) and the manifold hypothesis. Research in this area focuses on understanding the reasons why commonly-used DGMs succeed or fail at learning distributions supported on unknown low-dimensional manifolds, as well as developing new models explicitly designed to account for manifold-supported data. This manifold lens provides both clarity as to why some DGMs (e.g. diffusion models and some generative adversarial networks) empirically surpass others (e.g. likelihood-based models such as variational autoencoders, normalizing flows, or energy-based models) at sample generation, and guidance for devising more performant DGMs. We carry out the first survey of DGMs viewed through this lens, making two novel contributions along the way. First, we formally establish that numerical instability of high-dimensional likelihoods is unavoidable when modelling low-dimensional data. We then show that DGMs on learned representations of autoencoders can be interpreted as approximately minimizing Wasserstein distance: this result, which applies to latent diffusion models, helps justify their outstanding empirical results. The manifold lens provides a rich perspective from which to understand DGMs, which we aim to make more accessible and widespread.
Submission history
Access paper:.
- Other Formats
References & Citations
- Google Scholar
- Semantic Scholar
BibTeX formatted citation
Bibliographic and Citation Tools
Code, data and media associated with this article, recommenders and search tools.
- Institution
arXivLabs: experimental projects with community collaborators
arXivLabs is a framework that allows collaborators to develop and share new arXiv features directly on our website.
Both individuals and organizations that work with arXivLabs have embraced and accepted our values of openness, community, excellence, and user data privacy. arXiv is committed to these values and only works with partners that adhere to them.
Have an idea for a project that will add value for arXiv's community? Learn more about arXivLabs .
Deep-kidney: an effective deep learning framework for chronic kidney disease prediction
- Open access
- Published: 01 December 2023
- Volume 12 , article number 3 , ( 2024 )
Cite this article
You have full access to this open access article
- Dina Saif 1 ,
- Amany M. Sarhan 1 &
- Nada M. Elshennawy 1
2040 Accesses
Explore all metrics
Chronic kidney disease (CKD) is one of today’s most serious illnesses. Because this disease usually does not manifest itself until the kidney is severely damaged, early detection saves many people’s lives. Therefore, the contribution of the current paper is proposing three predictive models to predict CKD possible occurrence within 6 or 12 months before disease existence namely; convolutional neural network (CNN), long short-term memory (LSTM) model, and deep ensemble model. The deep ensemble model fuses three base deep learning classifiers (CNN, LSTM, and LSTM-BLSTM) using majority voting technique. To evaluate the performance of the proposed models, several experiments were conducted on two different public datasets. Among the predictive models and the reached results, the deep ensemble model is superior to all the other models, with an accuracy of 0.993 and 0.992 for the 6-month data and 12-month data predictions, respectively.
Similar content being viewed by others

Prediction of Chronic Kidney Diseases Using Deep Artificial Neural Network Technique

A Study on Machine Learning and Deep Learning Techniques Applied in Predicting Chronic Kidney Diseases

Performance Analysis of Machine Learning Algorithms in the Systematic Prediction of Chronic Kidney Disease on an Imbalanced Dataset
Avoid common mistakes on your manuscript.
Introduction
Chronic kidney disease (CKD) refers to kidney damage caused by an inability to filter blood properly. The kidneys’ primary function is to filter extra water and waste from human blood and then excrete them through urine. That is, when a person has CKD, wastes accumulate in his body and cause symptoms such as back pain, diarrhea, nosebleeds, fever, rash, vomiting, and abdominal pain. As a result of the damage occurring gradually over time, it will affect the rest of the human body and lead to the emergence of many diseases. As the disease advances and reaches its final stages, it may result in death.
Because of the lack of early detection of these diseases, mortality from infection with many diseases has recently increased. As a consequence, many studies have emerged to address this issue, assist doctors, and reduce mortality through the use of advanced computer-based detection techniques. Early diabetes prediction, for example, has been proposed using the Random Forest and XGBoost algorithms [ 1 ]. Furthermore, for disease detection, a multi-layer perceptron and Random Forest algorithms were used [ 2 , 3 ]. For predicting the likelihood of a human being a recent or future heart disease patient, a convolutional neural network model was developed [ 4 ]. For the prediction of coronary heart disease, an accuracy-based-weighted aging classifier ensemble algorithm was proposed [ 5 ].
CKD prediction using deep learning techniques, which is our main interest in this paper, is a very significant application of intellectual intelligent systems because it predicts the disease before it occurs, which greatly contributes to saving people’s lives. As an outcome, in order to defeat this dangerous disease, an effective mechanism for predicting CKD must be developed. Some studies focus on early detection, while only a few focus on predicting disease before it occurs. Multiple studies used Support Vector Machine and Artificial Neural Networks, Deep neural networks, an Ensemble algorithm, Extra tree, Random Forest, and Logistic Regression models to detect CKD at an early stage [ 6 , 7 , 8 , 9 , 10 , 11 ]. Furthermore, for CKD classification, the Density-based Feature Selection (DFS) with Ant Colony based Optimization (D-ACO) algorithm was proposed [ 12 ]. In terms of CKD prediction, Decision tree, Random forest, LightGBM, Logistic Regression, and CNN models have been developed to predict the disease 6–12 months in advance [ 13 ].
According to a massive amount of research in the machine learning field, two algorithms currently dominate this field: Ensemble and Deep Learning algorithms [ 14 ]. Deep learning is the gold standard of machine learning algorithms, and deep ensemble algorithms are a catch-all term for approaches that combine multiple deep learning classifiers to make a decision. As a result, in the current work, we use an ensemble algorithm in conjunction with ensemble and deep learning approaches. Deep learning techniques, on the other hand, are regarded as the most dominant and powerful player in a variety of machine learning challenges. It is primarily responsible for making a final prediction by locating hidden information in the massive dataset that doctors frequently obtain from patients. The deep learning model attempts to learn features that traditional techniques would not be able to extract. The use of this algorithm improves detection and prediction accuracy by avoiding the drawbacks of traditional learning techniques [ 15 ]. Deep learning techniques now outperform traditional classification techniques in terms of performance. Convolutional neural networks [ 16 ], long short-term memory [ 17 ], and many other techniques are used by deep learning algorithms to solve machine learning challenges. Over the last few years, many algorithms that combine ensemble algorithms and deep learning models have been developed in an attempt to improve predictive performance. The deep ensemble learning algorithm combines the benefits of both deep learning and ensemble learning to produce a final model with the best generalization performance [ 18 ].
In the case of kidney disease, scientists have attempted to detect it early or predict its occurrence. Disease detection implies that the patient already has the disease, whereas disease prediction implies that it will occur in the future. As a result, research in this area has been divided into two branches: detection and prediction. There are numerous studies in this field for the first type [ 13 ].
We faced the following challenges after reviewing the literature:
There isn’t enough data on CKD. The datasets for previous studies were based on medical tests. It does, however, consist of a small number of samples.
Previous research has focused on detecting the disease after it has occurred.
Due to a lack of data, the work in this field has not been fully explored.
There has only been one previous study that attempted to predict the disease in advance. However, the precision of this work was not high.
According to the previous challenges, the mortality rate of CDK disease is rapidly increasing.
As a result, in this paper, we will investigate several deep learning models as well as the ensemble approach to merge these models in order to fill a gap in the field. As a result, the following are the main contributions of this paper:
We propose two deep learning predictive models to predict CKD 6 or 12 months before disease occurrence, which are as follows:
Convolutional Neural Networks (CNN) Model.
Long Short-Term Memory (LSTM) Model.
We propose an ensemble model that uses the majority voting technique to combine three basic deep learning classifiers (CNN, LSTM, and LSTM-BLSTM) to improve classification performance.
For the task of CKD prediction, we train each model using two different public benchmark datasets. The first is to predict the disease 6 months ahead of time, while the second is to predict the disease 12 months ahead of time.
We assess the predictive models’ performance using various metrics to investigate the advantages and disadvantages of each. To demonstrate the strength of the proposed models, the results are compared to the results of the state-of-the-art work [ 13 ] using the same datasets.
In addition to the current section, section “Background and related work” presents previously developed approaches in risk detection and prediction for CKD, and classification ensemble techniques. The dataset is presented in section “ Materials and methodologies ” and the proposed models in described in detail. Section “ Proposed models evaluation ” evaluates the proposed predictive models, draws a comparative analysis, and discusses the prediction results. Section “ Conclusion ” concludes this paper.
Background and related work
Many researchers proposed algorithms for health risk prediction for a variety of diseases in an effort to reduce mortality. Li et al. [ 19 ] forecasted the risk of hospital readmission for diabetic patients using a combination of collaborative filtering-enhanced and deep learning approaches in 2018. With 88.94% accuracy, the algorithm outperforms the Naïve Bayes, SVM, and decision tree algorithms. Later that year, Alam et al. [ 3 ] created a medical data classifier based on the Random Forest algorithm and feature ranking for ten different diseases. The proposed method was based on determining the significance of features for classification using various feature ranking techniques.
In 2020, Bikku et al. [ 2 ] proposed a multi-layer perceptron algorithm based on supervised learning methods to predict the risk of various diseases (breast cancer, diabetes, and heart disease) with a high degree of certainty. Following that, Shankar et al. [ 4 ] developed a new technique based on Naïve Bayes and KNN to predict the likelihood of a human being a recent or future heart disease patient. The prediction of coronary heart disease entered the competition as well, with the introduction of an accuracy-based-weighted aging classifier ensemble algorithm (AB-WAE) [ 5 ]. On two different datasets, this algorithm achieved 93% and 91% accuracy.
Since diabetes classification has occupied researchers’ minds for the riskiness of this disease, the Random Forest and XGBoost algorithms have been applied to the PIMA diabetes dataset for early prediction of this disease. XGBoost algorithm was superior to Random Forest by achieving 74.10% accuracy, while Random Forest achieved 71% accuracy [ 1 ]. Random Forest was proven to be superior to XGBOOST in CKD prediction reaching an accuracy of 100% as in [ 9 , 11 ] using the CKD dataset [ 20 ].
Risk detection and prediction for chronic kidney disease
Given the riskiness of kidney disease to human health, scientists have attempted to detect it early or predict its occurrence in advance. Disease detection implies that the patient already has the disease, whereas disease prediction implies that it will occur in the future. As a consequence, research has been divided into two types: detection and prediction. In aspects of the first type, most of them used the same dataset [ 20 ], beginning with CDK detection. Almansour et al. [ 6 ] used SVM and ANN to detect CKD at an early stage. The dataset was preprocessed, and then the missing values were replaced. Following that, ten fold cross-validation was used. This study concluded that ANN outperformed SVM in terms of accuracy, with accuracy up to 99.75%. The limitation of this study is that the number of samples is limited, which causes the problem of dimensionality. This problem was solved by employing the SVM algorithm. This study suggests that a deep learning technique be used to detect CKD.
Elhoseny et al. [ 12 ] developed an intelligent classification technique for CKD in the same year, called Density-based Feature Selection (DFS) with Ant Colony based Optimization (D-ACO). This technique solved the problem of increasing the number of features in medical data by removing redundant features, which greatly aided in the resolution of many issues such as low interoperability, high computation, and overfitting. Using this method, the author achieved 95% detection accuracy with only 14 of the 24 features.
During the same year, Kriplani et al. [ 7 ] proposed a deep neural network model to detect the absence or presence of CKD in its early stages. This model used cross-validation to avoid overfitting and outperformed other available techniques, reaching 97% accuracy when compared to Naïve Bayes, Logistic, Random forest, Adaboost, and SVM.
Following that, in 2020, Jongbo et al. [ 8 ] used an ensemble algorithm: Random Subspace and Bagging to achieve 100% accuracy on the previous dataset, which is appropriate for efficient CKD diagnosis. The data is preprocessed, then missing values are handled, and finally the data is normalized. This algorithm was based on majority voting between three base-learners: KNN, Naïve Bayes, and Decision Tree. This study demonstrated that combining the base classifiers improved classification performance. In the performance matrices, the proposed model outperformed individual classifiers, according to the experimental results. In most cases, the random subspace algorithm outperformed the bagging algorithm.
During the same year, Ekanayake et al. [ 9 ] proposed an efficient method for detecting CKD based on medical data, beginning with data prepossessing and then filling missing values with K Nearest Neighbors-imputer, which resulted in higher detection model accuracy. Finally, the classification method was used. They focused on the practical aspects of data collection, emphasizing the importance of combining domain knowledge in CKD detection using machine learning. Among the 11 classifiers tested, the authors demonstrated the superiority of extra tree and Random Forest classifiers in CKD detection: (logistic regression, KNN, SVC with a linear kernel, SVC with RBF kernel, Gaussian NB, decision tree classifier, XGB classifier, Adaboost classifier, and a classical neural network). The K-Nearest Neighbors-imputer is recommended for this study to handle missing values in other diseases. Furthermore, adding more features to the analysis is important, such as food types, water consumption, and genomics knowledge.
In addition, Gudeti et al. [ 10 ] distinguished the performance of several machine learning techniques in 2020 based on their accuracy in analyzing CKD and distinguishing between CKD and Non-CKD patients. To detect CKD, the authors used Logistic Regression, SVM, and KNN models. The SVM model outperformed the other techniques, achieving 99.2% accuracy. The main benefit of this research is that the detection process is quick, allowing doctors to start treating patients sooner and further categorizing the patient population in less time. They did, however, use a small dataset of 400 patients.
Later that year, in 2021, Chittora et al. [ 21 ] detected CKD using full or important features. Full features, correlation-based feature selection, Wrapper technique feature selection, least absolute shrinkage and selection operator regression LASSO, synthetic minority over-sampling technique with least absolute shrinkage and selection operator regression selected features, and synthetic minority over-sampling method using full features were used to calculate the results. C5.0, CHAID, ANN, linear support vector machine (LSVM), logistic regression (LR), random tree (RT), and KNN were also used as classifiers. Finally, with the full features in synthetic minority over-sampling technique, LSVM achieved the highest accuracy of 98.86%.
Following that, Senan et al. [ 11 ] used machine learning techniques to develop a diagnosis system to detect CKD to aid experts in early diagnosis. The mean and mode were used to replace the missing values. Recursive Feature Elimination was used to select the most important features (RFE). The dataset was divided into two parts: 75% for training and 25% for testing and validation. Following that, four machine learning algorithms were used: support vector machine (SVM), Random Forest (RF), k-nearest neighbors (KNN), and decision tree (DT). To achieve the best results, the parameters were tuned for all classifiers. Among these four classifiers, the Random Forest (RF) algorithm outperformed all other four techniques by achieving 100% accuracy.
Finally, Singh et al. [ 22 ] proposed a deep neural network in 2022. The missing values were replaced by the average of the associated feature, and the Recursive Feature Elimination (RFE) algorithm was used to select features. The key parameters in the study were Specific Gravity, Hemoglobin, Red Blood Cell Count, Creatinine Levels, Packed Cell Volume, Albumin, and Hypertension (RFE). Following that, the selected features were fed into five classifiers (Deep neural network DNN, Naïve Bayes classifier, KNN, Random Forest, and Logistic regression). DNN outperformed all other models in terms of accuracy. The size of the dataset is a limitation of both the proposed algorithm and previous studies. The next step in this research will be to collect more sophisticated and representative CKD data to detect disease severity. The authors intend to use the proposed model on medical data containing the night urination, acid–base parameters, inorganic phosphorus concentration, and hyperparathyroidism features.
Concerning the second type of investigation, disease risk prediction, the first pioneering technique was proposed in 2021, which concerned CKD prediction as opposed to previous searches, which concerned CKD detection [ 13 ]. The primary goal of this study was to forecast the occurrence of CKD 6–12 months before disease onset using Taiwan’s National Health Insurance dataset [ 23 ]. The predictive model was developed using comorbidity, demographic, and medication data from patients over a 2-year period. For 12-month and 6-month predictions, the CNN model had the best AUROC of 0.954 and 0.957, with accuracy of 88% and 89%, respectively. While, the most important predictors were: gout, diabetes mellitus, age and medications such as angiotensin and sulfonamides. Table 1 summarizes the recent health risk prediction research for CKD.
Classification ensemble techniques
Ensemble techniques are considered state-of-the-art methodologies for solving problems in a wide range of machine learning applications. The intuitive motivation for ensemble stems from human nature and the proclivity to gather disparate viewpoints and integrate them in order to make a complex decision. This idea depends on integrating multiple base-learners to obtain a classifier that outperforms them all using one of the combination algorithms: [Average Ensemble (AE), Weighted Average Ensemble (WAE), and Majority Voting Ensemble (MVE)]. In recent years, machine learning researchers have demonstrated through hands-on experimental research that combining the outputs of multiple classifiers improves the performance of a single classifier [ 18 ]. The ensemble technique has been used in a variety of applications, including disease detection and prediction, due to its impact on several machine learning challenges [ 5 , 8 , 24 , 25 ]. The ensemble technique’s main idea is to maximize predictive performance by combining the strengths of multiple individual classifiers. In other words, the goal of deep ensemble models is to create a model that incorporates the advantages of both ensemble and deep models.
Recently, there have been some issues with using an individual classifier, such as overfitting, class imbalance, concept drift, and the curse of dimensionality, which cause a single classifier prediction to fail [ 26 ]. As a result, this new method has emerged in scientific research to address these issues. The predictive accuracy improves by using this algorithm in different machine learning challenges. The main idea of any ensemble learning is to use a combination function F to combine a set k of individual classifiers, c 1 , c 2 , …, c k , to predict a single output. Given a dataset of size n and features of dimension m, D = {( x , y )}, 1 ≤ i ≤ n, x i ∈ R m , the output’s prediction of this method is shown in Eq. ( 1 ) [ 27 ].
In this section, we will examine the most common Ensemble techniques that are commonly used in many machine learning applications, as well as some literature reviews on using ensemble techniques in health risk prediction.
Average ensemble (AE)
This technique demonstrated its high efficiency in scientific research. The main idea behind the techniques is that the final prediction is calculated by taking the average of the individual learners’ outputs. This average is calculated directly from the outcomes of individual learners or by applying the softmax function to the forecasting probabilities of the classes, as shown in Eq. ( 2 ). The performance of this technique is improved because of variance among the models is reduced [ 18 ].
where P i J is the probability of the outcome of the ith unit on the jth base learner, \({O}_{j}^{i}\) is the output of the ith unit of the jth base learner and K is the number of the classes. This approach is appropriate when individual performance is proportional [ 28 ]. On the other hand, it is not appropriate when individual classifier performance is grossly disproportionate. The overall performance will be reduced in this case due to the influence of weak learners.
Because this technique does not take into account the performance of individual models, all models have the same weight. The previous method has the limitation that the results of the weak base classifier will have an adverse effect on the final model output. To avoid this problem, the Weighted Average Ensemble (WAE) is proposed, which provides sorted weights for models based on their efficiency.
Weighted average ensemble (WAE)
The previous approach is appropriate when the performance of the individuals are proportional [ 28 ]. On the other hand, it isn’t appropriate when the individual learners’ performances are absolutely disproportionate. In this case, the overall performance will be reduced according to the influence of weak learners. To avoid this problem, the (WAE) is proposed, which provides sorted weights for models based on their efficiency. It is thought to be an extension of the previous method, in which the final prediction value is obtained by calculating the average of all the base classifiers’ predictions. In contrast, in the weighted average, each data point is assigned a pre-defined weight to indicate its importance in the prediction, and the final prediction value is calculated by taking the weighted average of all the data points into account. Each classifier in the ensemble contributes to the final prediction based on its weight in this technique. The final prediction for class label prediction is calculated using the mode of the individuals’ predictions. For the class probability prediction, the final prediction is calculated using argmax of the summed probabilities of each class label [ 29 ].
Majority voting ensemble (MVE)
In the research field, this technique is regarded as the most widely used approach in the ensemble technique. This technique, similar to the previous ones, combines the outputs of individual learners. Instead of calculating the average of the probability results, (MVE) counts the votes of individual classifiers and predicts the final class based on the majority of votes [ 18 ]. The main advantage of this technique is that it is less biased towards the outcome of a specific individual learner because the majority vote count relieves the influence; additionally, the influence of weak learners is no longer significant. The majority voting rule comes in three varieties:
Unanimous voting, in which all individual classifiers agree on the prediction;
Simple majority voting, in which the prediction must be supported by at least 51% of all classifiers; and
Majority or plurality voting, in which individual learners’ votes are counted and the final prediction is calculated based on the majority of votes. The majority voting rule improves prediction performance.
This model caught the interest of scientists and researchers, and it is now used in a variety of applications in health risk detection and prediction for a variety of diseases.
Literature review of using ensemble in disease detection
This section examines the literature on ensemble learning in disease detection, with machine learning or deep learning as individual classifiers. Using ensemble learning, Raza et al. [ 30 ] created a model for detecting heart disease that is both reliable and accurate. The majority voting rule was used to combine the results of three classification algorithms [logistic regression (LR), multilayer perceptron (MLP), and Naïve Bayes (NB)] in this paper. The proposed ensemble method achieved classification accuracy of 88.88%, which is superior to any other base classifier.
Following that, Atallah et al. [ 24 ] presented an ensemble method based on the majority voting technique in the same field. This method combined Stochastic Gradient Descent (SGD), KNN, Random Forest, and Logistic Regression to provide doctors with greater dependability and accuracy. Finally, using the hard voting ensemble model, this technique achieved 90% accuracy. Yadav et al. [ 31 ] used various ensemble techniques on 10 biomedical datasets [ 32 ]. These techniques performed competitively against individual classifiers. The highest AUC was achieved using the average ensemble and the Rank Average Ensemble (RAE) in most datasets.
Individual classifiers were outperformed by these techniques. In most datasets, the average ensemble and the Rank Average Ensemble (RAE) produced the highest AUC. Similarly, Tao Zhou et al. [ 33 ] proposed an ensemble deep learning model called (EDL COVID) to detect COVID 19 in lung CT images, employing a relative majority vote algorithm with 99.05% accuracy. Before employing the Ensemble technique, the base models were built using ResNet, GoogleNet, and AlexNet. In terms of performance and detection speed, the EDL COVID classifier outperformed the single classifiers. Similarly, Chandra et al. [ 34 ] used the majority voting ensemble technique to create a two-phase classification system [normal vs. abnormal (phase-I) and COVID-19 vs. pneumonia (phase-II)]. The obtained precision for Phase-I and Phase-II was 98.062% and 91.329%, respectively.
Neloy et al. [ 25 ] proposed an ensemble model to achieve an excellent result in heart disease prediction. Among the baseline models used in their proposed work are (Random Forest, Decision Tree, and Naïve Bayes). The combining process, which used the Weighted Average Ensemble technique, achieved 100% accuracy on training and 93% accuracy on testing [ 25 ].
Using voice recordings of 50 patients and 50 healthy people, Hire et al. proposed an ensemble algorithm of CNNs for detecting Parkinson’s disease. The publicly available database obtained from PC-GITA was used. The base classifier was trained using a multiple-fine-tuning method. Each vowel was trained and tested separately, then a tenfold validation was performed to test the models. The proposed approach was soundly able to differentiate between the voices of patients and the healthy people for all vowels. The proposed model achieved 99% accuracy, 93.3% specificity, 86.2% sensitivity, and 89.6% AUC. The monitoring of the patients can be applied online without needing additional hardware.
Table 2 summarizes the ensemble disease detection techniques, the dataset used in the experiments, and the highest accuracy.
Materials and methodologies
Dataset description.
The datasets publicly released from Taiwan’s National Health Insurance Research Database (NHIRD) [ 39 ] is used in our study. The author of [ 13 ] compiled patient data into CSV files in order to predict CKD disease 6–12 months in advance [ 23 ]. Each patient’s data was saved for 2 years, and it consisted of 965 comorbidities denoted by ICD 9 codes (International Classification of Diseases) and 537 medications denoted by Anatomical Therapeutic Chemical code (ATC codes) for 6 months’ data. It includes 967 comorbidities and 539 medications for a 12-month period of data, as well as the patient’s age and gender. There is also a class label that represents CKD. Each patient is labelled with a binary number (0 means the patient will not get CKD after the specified period, while 1 means he will get CKD after a certain period). A total of 90,000 patients are analyzed, divided into 18,000 with CKD and 72,000 without CKD diagnose separately for each dataset.
Figure 1 represents a part of a sample of the used dataset [ 39 ]. As we see in the figure, “ckd” represents the target column, while “age” represents the patient’s age. The “sex” column denotes the patient’s gender, while (1–5) represents the “ICD_9” code, which represents a number of diseases, including Cholera disease, Typhoid and paratyphoid fevers, Salmonella infections, Shigellosis, and poisoning. For each patient, a zero indicates that he was not infected with the disease throughout the observation period of 2 years; otherwise, it indicates that he was infected.
Part of a sample of the used dataset
Methodologies
The prediction problem is treated as a classification problem, with the model’s output being either 0 or 1. (0 means the patient will not get CKD after the specified period, while 1 means he will get CKD after a certain period). We present the architecture of the three proposed predictive models for chronic kidney disease prediction in this section (CKD). Because there has only been one work directed toward solving this problem [ 13 ], we intend to explore different models for the problem using deep learning models in our presented work. Deep learning algorithms are the gold standard in machine learning. It is useful in a variety of applications when analyzing large amounts of data. 90,000 patients were analyzed in our two datasets, with 1504 features for 6 months’ data and 1508 features for 12 months’ data. As a result, this algorithm can predict CDKs by uncovering hidden information in the large dataset that doctors frequently obtain from patients. The deep learning model attempts to learn features that traditional techniques cannot extract. Using the same datasets, traditional machine learning techniques did not achieve the desired accuracy. As a result, the use of this algorithm improves detection and prediction accuracy by avoiding the drawbacks of traditional learning techniques. The first two predictive models are deep learning models: CNN and LSTM, while the third is an ensemble model composed of three different deep learning models.
Convolutional neural networks (CNNs) and long short-term memory networks (LSTMs) were chosen in the context of CKD prediction due to their unique advantages. When working with tabular data that may contain spatially important information in the context of CKD, it is essential to be able to capture spatial patterns, which CNNs are good at doing. However, LSTMs are excellent at modelling temporal relationships, which makes them a good choice for examining sequential data from the CSV file that can capture the development of CKD-related traits over time. A comprehensive approach to feature extraction that takes into account the dataset’s spatial and temporal dimensions is made possible by combining CNNs with LSTMs. We use these models to enhance prediction accuracy by capturing comprehensive patterns and relationships within the data.
The methodologies adopted in this work are depicted schematically in Fig. 2 . Each model is trained twice for different prediction tasks (the first time using 6 months of data and the second time using 12 months of data). Each model’s input is a CSV file containing 90,000 samples with 1504 features for 6 months’ data and 1508 features for 12 months’ data. The input features have been reshaped before applying the model to match the model requirements, while the output is a binary number that represents the class. The same model structures are used for both benchmark datasets, but the input layer is reshaped differently due to the difference in the number of features in each.
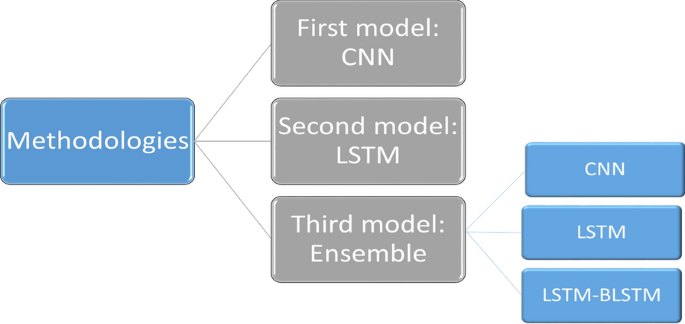
Schematic diagram for the methodologies
Convolutional neural network (CNN)-CDK predictive model
CNN-based models have demonstrated robust performance in a variety of research applications. As a result, the first proposed predictive model in our work, as illustrated in Fig. 3 and Table 3 , is based on this robust model. The input layer receives 1504 features for 6 months’ data, which are then reshaped to (47 × 8 × 4). While the input layer receives 1508 features for a 12-month period data, which is then reshaped to (29 × 13 × 4) to match the network configuration. The output layer is made up of a neuron that determines the class (1 representing CKD and 0 representing Non-CKD).
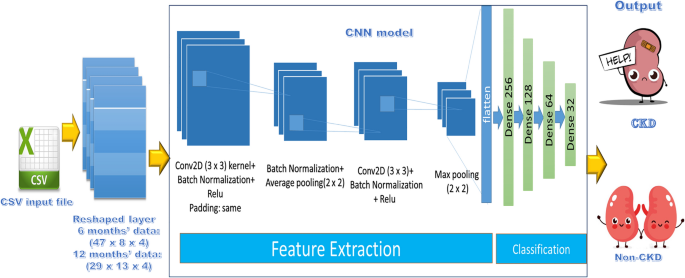
Proposed CNN predictive model for CKD prediction
The activation function is applied to obtain the output as shown in Eq. ( 3 )
where h k represents the output feature map, \(x\) is the input, b k and W k are the bias and the weight of the k neuron, respectively [ 40 ].
The average pooling technique has been adopted, with a kernel size of 2. This technique works by moving a pool with a specific size over the incoming input, then calculating the average value in each one. Moreover, the max pooling technique has been utilized, with a kernel size of 2. Furthermore, the dropout mechanism is used to avoid overfitting and improve network performance.
Long short-term memory (LSTM)-CDK predictive model
LSTM is a type of deep learning network model that is frequently used in time-series signals analysis. The most significant advantages of this model are [ 41 ]: it has a higher accuracy in long-term dependency problems than Recurrent Neural Network (RNN). Furthermore, vanishing gradients problems can be solved using memory blocks using this technique.
The LSTM unit consists of an input gate I t , an output gate O t and a forget gate F t . The three gates’ activations are computed using the subsequent equations [ 42 ]:
The sigmoid activation function and the current input are represented as σ and X t respectively. The input weights are denoted as W i , W f and W o while b i , b f and b o are the bias. Whilst the recurrent weights are symbolized as R i , R f and R o . The output of the previous block is represented as H t−1 . The modified new memory \(\overline{C }\) t is computed as in Eq. ( 7 ):
where tanh(·) represents the hyperbolic tangent function. Whilst R t and W t denote the recurrent weight and input weight respectively. The computation of the current memory cell C t is illustrated as in Eq. ( 8 ):
where C t−1 represents the previous memory cell while ⊙ indicates the element-wise multiplication operation. To calculate the LSTM output H t , the following equation is used:
The input layer receives 1504 features for 6 months’ data, then reshaped to (47 × 32). While it is reshaping to (52 × 29) for 12 months’ data to match the network configuration. The structure diagram and the architecture of LSTM are illustrated in Fig. 4 and Table 4 .
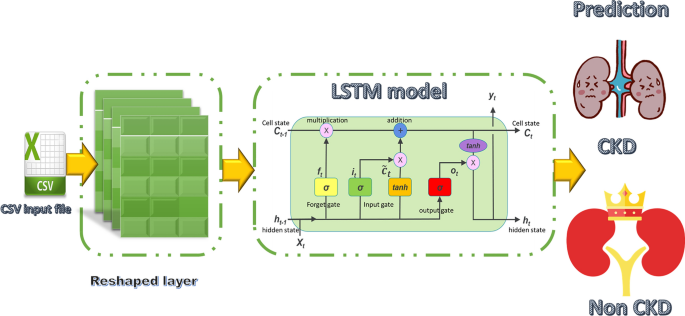
Proposed LSTM predictive model for CKD prediction
Deep ensemble predictive model (DEM)
Ensemble learning methods are usually used to improve prediction performance when a single classifier is insufficient to achieve a high level of performance. The main idea behind this predictive model is to aggregate a group of different individual classifiers to improve performance by combining a weak classifier with a strong classifier to increase the efficiency of the weak learner. In our proposed ensemble model, we combine CNN, LSTM, and LSTM-BLSTM models to produce an effective computational model for CKD prediction based on majority voting ensemble, as shown in Fig. 5 . The majority voting ensemble was chosen due to its robustness and because it is less biased towards the outcome of a particular individual learner. Furthermore, the influence of weak learners is no longer significant, and finally, its impressive results in disease detection are documented in the literature [ 18 , 24 , 25 , 30 , 31 , 33 , 34 ].
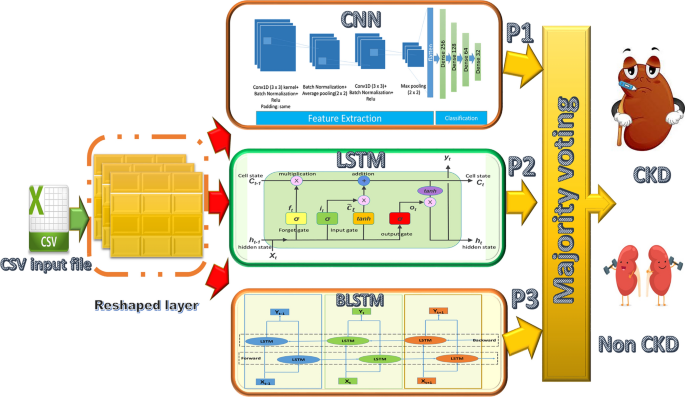
Structure of the proposed ensemble CDK predictive model
The following subsections provide details on each individual model in the ensemble model.
First model in the ensemble: CNN model_2
The structure of the ensemble’s first model is a CNN model, referred to as CNN model_2, as depicted in Fig. 5 and Table 5 . The figure illustrates the application of a 1D CNN in our proposed model to generate a fast, generic, and highly accurate CKD predictive model. The 1D convolution is represented by the following equation:
where \({b}_{k}^{l}\) is the bias for layer l of the kth neuron, \({x}_{k}^{l}\) is the input for the same layer, \({s}_{i}^{l-1}\) is the output of the ith neuron at layer l − 1, \({w}_{ik}^{l-1}\) is the kernel (filter) from layer l − 1 to layer l.
The output, \({y}_{k}^{l}\) , can be calculated by passing the input \({x}_{k}^{l}\) through the activation function as follows:
The back-propagation algorithm (BP) is then used to reduce the output error. This algorithm works its way backwards from the output layer to the input layer. Consider the output layer (L). The number of classes is represented by NL, and for an input vector p, the target and output vectors are represented by \({t}_{i}^{p}\) and [ \({y}_{1}^{L}\) ,⋯, \({y}_{NL}^{L}\) ], respectively. As a result, the mean-squared error (MSE), E p , can be computed as follows:
The derivation is used, and the various gradients of the neurons are computed recursively. As a result, the network’s weights are updated until the least error is reached.
This model is composed of Conv1D, avg_pooling, Conv1D, max_pooling, dropout, flatten, dense 256, dense 128, dropout, dense 64, dropout, dense 32 which is finally connected to another dense layer for CKD prediction.
Second model in the ensemble: LSTM model_2
We use LSTM in our model to avoid the vanishing gradient problem and to build a high-performance computational framework predictive model. The same equations as mentioned in section “ Deep ensemble predictive model (DEM) ” are used. The model is made up of an LSTM layer with 500 hidden units. Then, another LSTM layer with 200 hidden units is added. The previous layers are followed by a dense layer of 128 neurons. A dropout is used, followed by a second dense layer of 64 neurons. The dropout is used again to avoid overfitting and improve model performance. Following these layers is a dense layer of thirty-two neurons, which is finally connected to another dense layer for CKD prediction.
Third model in the ensemble: LSTM-BLSTM model
As shown in Fig. 5 and Table 5 , the third model in the ensemble is a hybrid model that combines LSTM and BLSTM in an attempt to improve the performance of the ensemble models. A Bidirectional LSTM (BLSTM) is an enhanced version of LSTM. It is made up of two LSTMs that work in opposite directions (forward and backward). The forward direction is represented by \({h}_{t}^{f}\) that denotes the input in ascending order, i.e., t = 1, 2, 3, …, T. The opposite direction is represented by a backward hidden layer called \({h}_{t}^{b}\) , which represents the input in descending order, i.e., t = T, …, 3, 2, 1. Finally, \({y}_{t}\) is generated by combining \({h}_{t}^{f}\) and \({h}_{t}^{b}\) . The BLSTM model is represented by the following equations:
where W is a weight matrix ( \({W}_{xh}^{f}\) is a weight that connects input (x) to the hidden layer (h) in the forward direction, while \({W}_{xh}^{b}\) is the same but in the backward direction). \({b}_{h}^{f}\) is a forward direction bias vector, whereas, \({b}_{h}^{b}\) is a backward direction bias vector, the out is symbolized by \({y}_{t}\) [ 43 ].
Proposed models evaluation
The experiments are carried out using a publicly available dataset [ 23 ] that contains two different types of samples. The first one represents CKD prediction over 6 months earlier, while the other represents CKD prediction over 12 months earlier. Each one involves 90,000 samples, with 80% of them used for training and 20% for validation. The aforementioned models were implemented using Python 3 involving the Keras framework running on Google Colab using a GPU on processor: (Intel(R) Xeon(R) CPU @ 2.20 GHz) with 13 GB RAM.
The classification process used by the trained deep learning models is applied on the test dataset. As for the first two models (CNN and LSTM) models, the new sample is fed to each model to generate the final prediction. On the other hand, when a test sample is fed to the proposed Ensemble model, it is first distributed to all individual models. Next, each classifier produces a prediction. After that, the majority voting technique is applied to all base classifiers’ results to generate the final prediction.
Performance metrics
To compare the models’ performance, four commonly used performance evaluation metrics were used: true negative (TN), true positive (TP), false negative (FN), and false-positive (FP). Furthermore, five metrics are used in the evaluation: Recall, Precision, Accuracy, F1_score, and specificity which are calculated as given in Eqs. ( 16 )–( 20 ). A recall is the number of positive instances predicted from the total number of positive instances; it is also known as sensitivity or true positive rate. Precision, also known as Positive Predictive Value, is the number of instances predicted as positive out of the total number of samples predicted as positive. Accuracy is defined as the number of correctly predicted instances divided by the total number of instances. F1-score combines Precision and Recall into a single metric using their harmonic mean. The number of instances predicted as negative out of the total number of negative instances is referred to as specificity.
where TP denotes true positive or correctly classified positive class, TN denotes true negative or correctly classified negative class, FP denotes false positive or incorrectly classified positive class, and FN denotes false negative or incorrectly classified negative class.
To assess the impact of the proposed deep ensemble approach on prediction results, we ran several experiments on the aforementioned benchmark datasets and compared the ensemble’s performance to all individual models. Finally, we present all experimental results and compare them to previous recent results [ 13 ].
Experimental results and comparative analysis
This section presents the results of the proposed models, as well as the performance evaluation metrics mentioned in section “ Performance metrics ”. Tables 3 and 4 show the structure of the CNN and LSTM models used in this study. The activation function of the convolution layer and dense layer is (ReLU: Rectified Linear Unit), and the model employs the (Adamax) optimizer with a learning rate (lr) of 0.0009, with all other parameters set to default values.
Figure 5 depicts the structure of the ensemble model. This model is made up of three distinct models (CNN, LSTM, and LSTM-BLSTM). First, we present the individual classifiers’ classification results. The performance of the ensemble-based model is then evaluated. We adopted the majority voting ensemble (MVE) because it is the most widely used approach in the research area because it avoids the limitations of other techniques mentioned previously, and it also performs well in many approaches. The results of the proposed models are compared to each other. Furthermore, a comparison to the results of previous work [ 13 ] is made.
Tables 6 and 7 compare our proposed models to each other in order to assess their performance from various perspectives, whereas Tables 8 and 9 compare our work to the previous study [ 13 ] using the same metrics found in their paper. Tables 6 and 7 show an evaluation of all proposed models for 6 months and 12 months’ data in terms of Precision, Sensitivity, F1-score, Specificity, and Accuracy. These values are shown in the detailed results for CKD and Non-CKD separately, as well as the macro and weighted averages. Tables 8 and 9 show a comparison of the models proposed in this study and previous work [ 13 ] on the same datasets in terms of Precision, Sensitivity, Specificity, F1-score, and Accuracy. The values in bold font represent the best accuracy achieved in the compared models. These results show that the ensemble model outperforms the individual models and previous work results in many aspects: sensitivity, precision, specificity, F1-score and accuracy. The proposed model has proven its worthiness in all these respects.
Figures 6 and 7 display a graphical representation of the performance of all proposed models as well as the models in the comparative paper [ 13 ] on the same datasets for both 6 months and 12 months’ data. The figures show the model’s performance improvement over previous models.
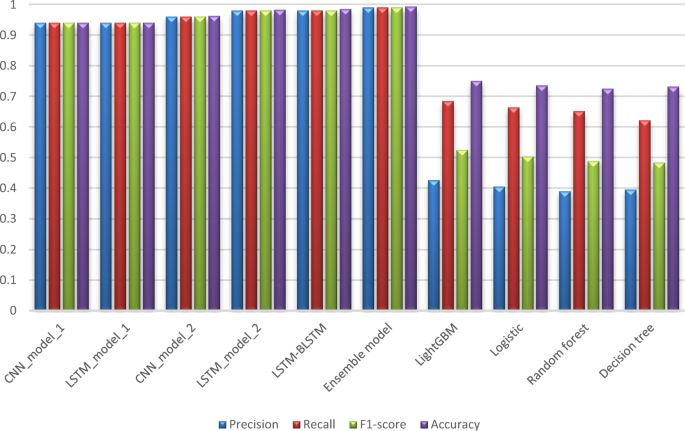
Comparison of performance metrics for 6-month data
Figures 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 and 19 show the confusion matrices for the predictive models, which represent the confusion matrix results for 6 months and 12 months’ data, respectively. The figures demonstrate the model’s classification robustness in the two statuses, CKD and Non-CKD. Finally, Figs. 18 and 19 show the ensemble model’s confusion matrix results. This model’s strength and outstanding performance in many types of research also demonstrate its worthiness in our case. Unfortunately, this model requires more memory space and takes longer than the individual models, indicating that its computational complexity is higher than the others. However, its accuracy outperforms that of the individual models.
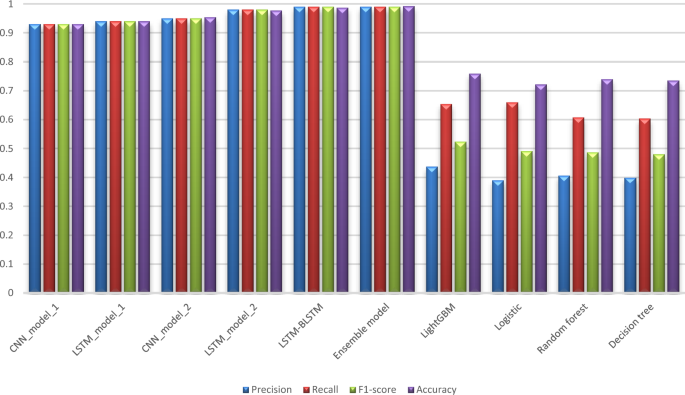
Comparison of performance metrics for 12-month data
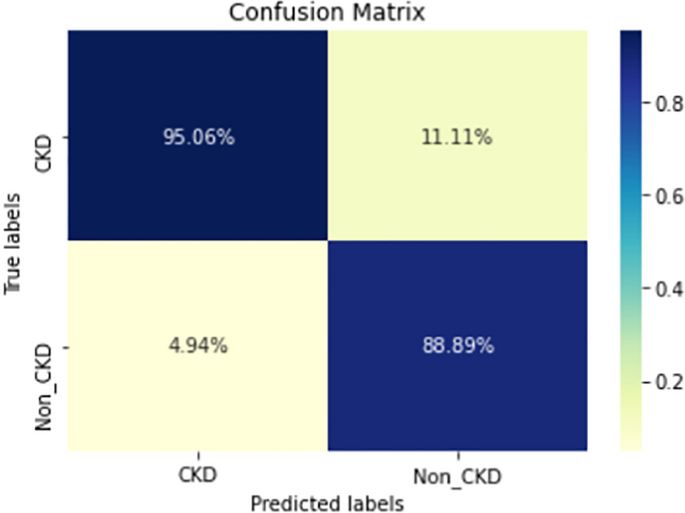
Confusion matrix of CNN model_1 (6_months data)
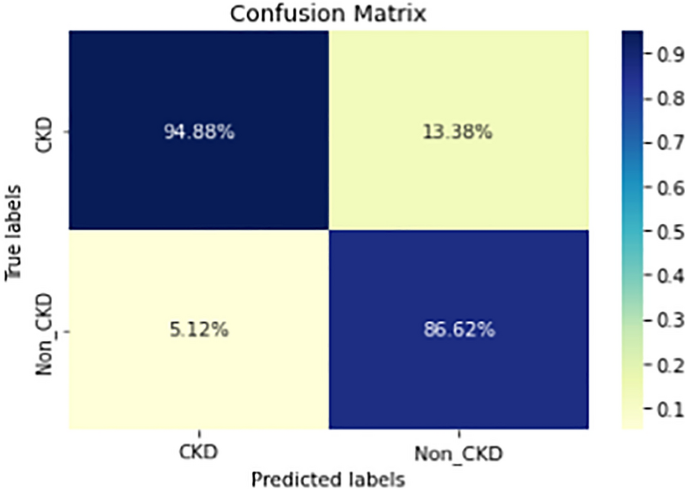
Confusion matrix of CNN model_1 (12_months data)
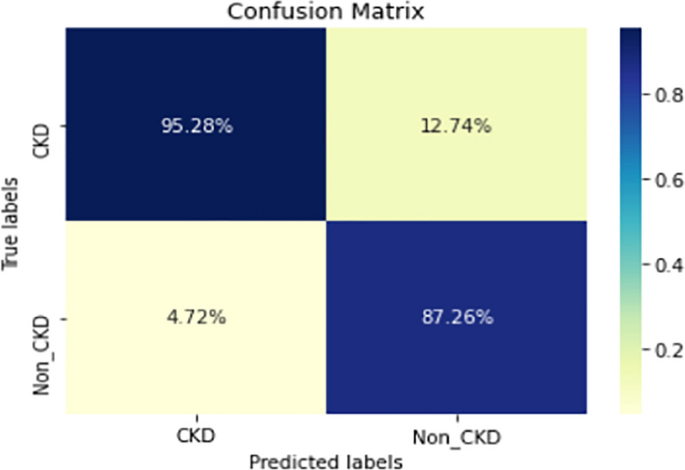
Confusion matrix of LSTM model_1 (6_months data)
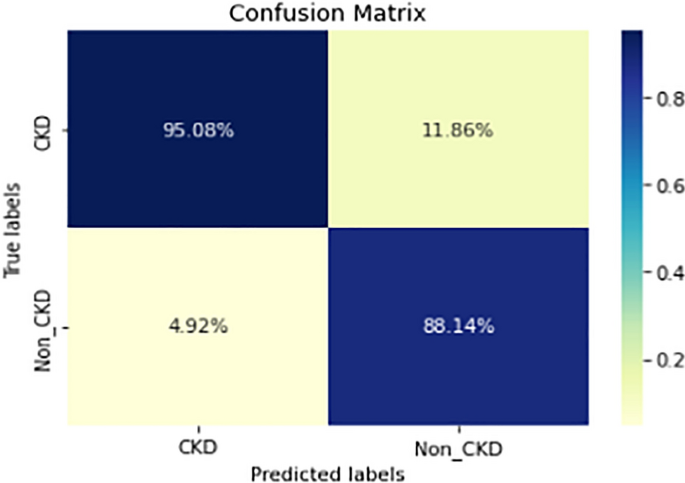
Confusion matrix of LSTM model_1 (12_months data)
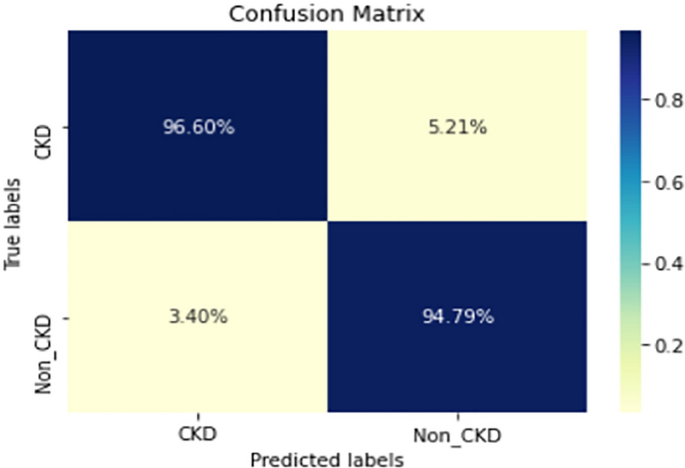
Confusion matrix of CNN model_2 (6_months data)
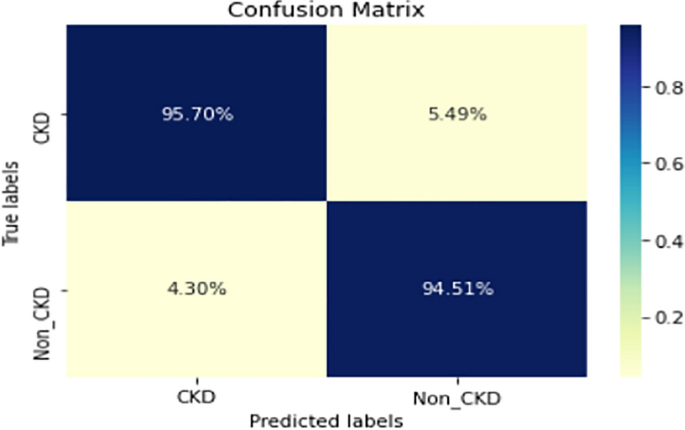
Confusion matrix of CNN model_2 (12_months data)
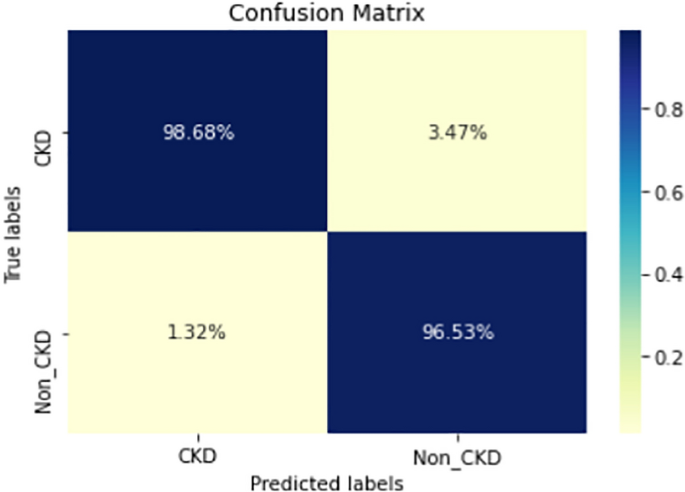
Confusion matrix of LSTM model_2 (6_months data)
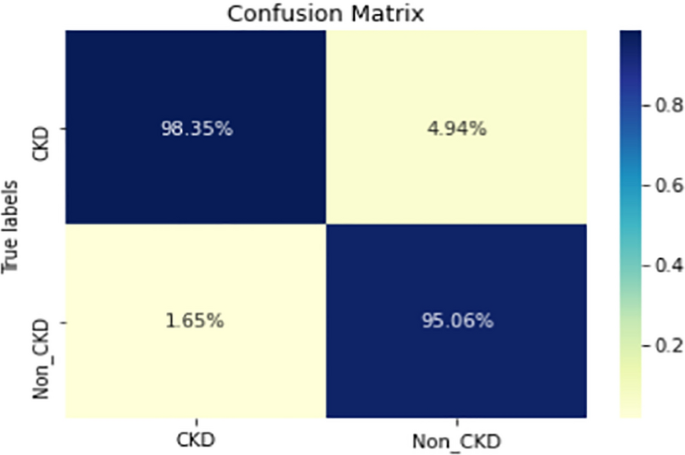
Confusion matrix of LSTM model_2 (12_months data)
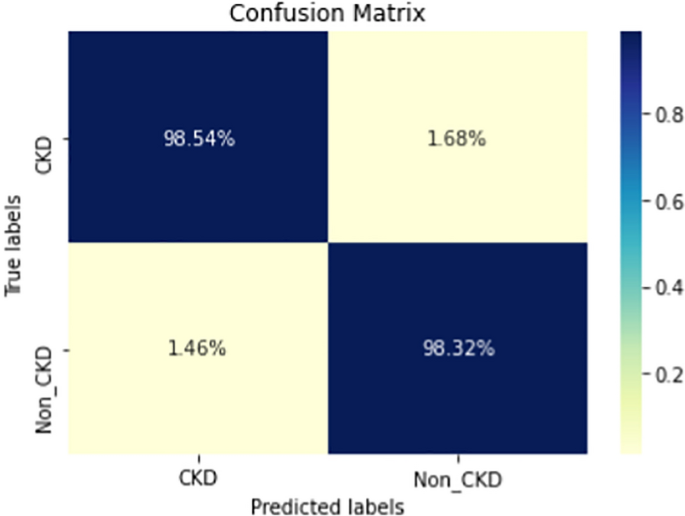
Confusion matrix of LSTM-BLSTM model (6_months data)
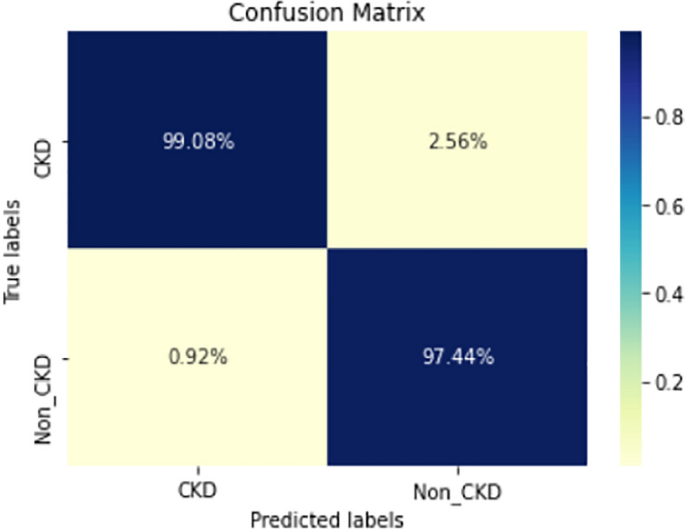
Confusion matrix of LSTM-BLSTM model (12_months data)
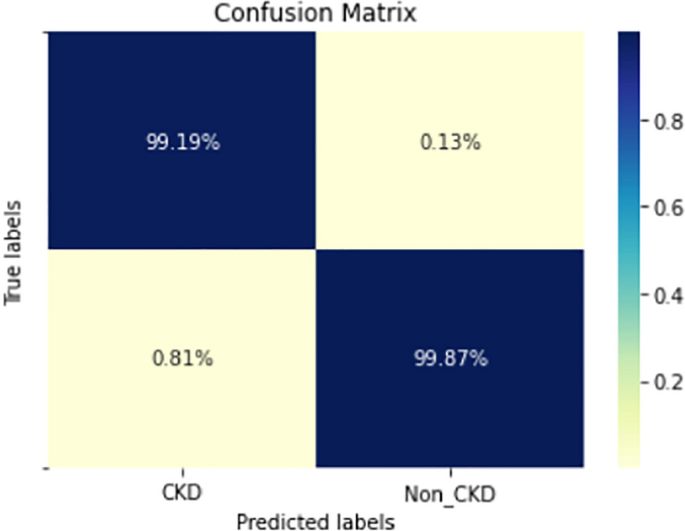
Confusion matrix of Ensemble model (6_months data)
Results and discussion
The main objective of the proposed research is the classification of CKD and Non-CKD classes with higher classification performance. Recently, the number of publicly available CKD datasets has become limited and contain only a small number of samples. Hence, the use of existing datasets with a large number of samples strongly supports the performance of the models. The main advantage of this method is that it does not need laboratory data as related studies in this field. However, it necessitates demographic information such as age and gender, in addition to prescriptions and diagnoses from patients. This information is widely available and easy to obtain from the appropriate authorities.
The experimental results show that the accuracy of the (LSTM_BLSTM) model outperforms all other individual deep learning models, as shown in Tables 6 and 7 for each individual model, with an accuracy score of 98.49%, 98.7% for 6 months and 12 months’ data, respectively. Furthermore, the confusion metrics show that the BLSTM-LSTM model is capable of classifying CKD and Non-CKD cases with 98.54%, 98.32% for 6 months’ data and 99.08%, 97.44% for 12 months’ data, respectively. The findings of the comparison show that the LSTM model outperforms the CNN model. For 6 months and 12 months’ data, the accuracy of the second CNN model increased by 2.26% and 2.4%, respectively, over the accuracy of the first CNN model. As we utilized conv_2D in the first CNN model and conv_1D in another, modifying the structure of the model enhances the outcomes of the base models and produced better results. Furthermore, we employ the LSTM model separately and integrate it with the BLSTM model to create a hybrid model, which also enhanced the results. Finally, combining the best models to create an ensemble model enhances the overall performance. It is also clear that using the ensemble approach leads to significant improvements in the model’s prediction capability, as the results show that the performance using the proposed ensemble model that combines (CNN, LSTM, and LSTM_BLSTM) models outperformed all other models significantly, with an accuracy score of 99.31% for 6 months, and 99.2% for 12 months, respectively. This is accomplished by strengthening the classification step with the ensemble algorithm’s majority voting capability, which increases prediction accuracy. As a result, our findings suggest that the proposed ensemble model has improved complex feature representations and will perform well in the classification task.
According to previous research [ 13 ], the LightGBM achieved the highest accuracy among the other machine learning techniques on the various aggregated datasets, with 0.751 and 0.759 for 6 months and 12 months’ data, respectively. Deep learning techniques, as shown in Tables 8 and 9 , significantly improved accuracy in our study. While using the ensemble method has greatly improved the results. Figures 6 and 7 demonstrate a summary of the comparison with literature-based approaches developed for CKD prediction using the same dataset. This method is appropriate for a population study but not for assisting clinicians with individual patients. To achieve the best results in patient diagnosis, decisions should be based on laboratory tests [ 13 ].
The following are the key features of the proposed research in CKD prediction:
Our proposed models, which are based on deep learning models, can effectively predict CKD 6 or 12 months before its occurrence, which is considered a scientific breakthrough and contributes to saving people’s lives.
Deep learning techniques outperform traditional machine learning approaches in terms of performance because of their ability to extract features from input data without the intervention of a human. As a result, a better classification model is produced.
If the model parameters are well designed and optimized, changing the structure of the CNN and LSTM models leads to better results.
A hybrid model (LSTM_BLSTM) outperforms all other individual deep learning models with an accuracy score of 98.49%, for 6 months, and 98.7% for 12 months, which is higher than the existing recent models.
When compared to traditional machine learning approaches and individual deep learning models, using an ensemble model can significantly improve accuracy.
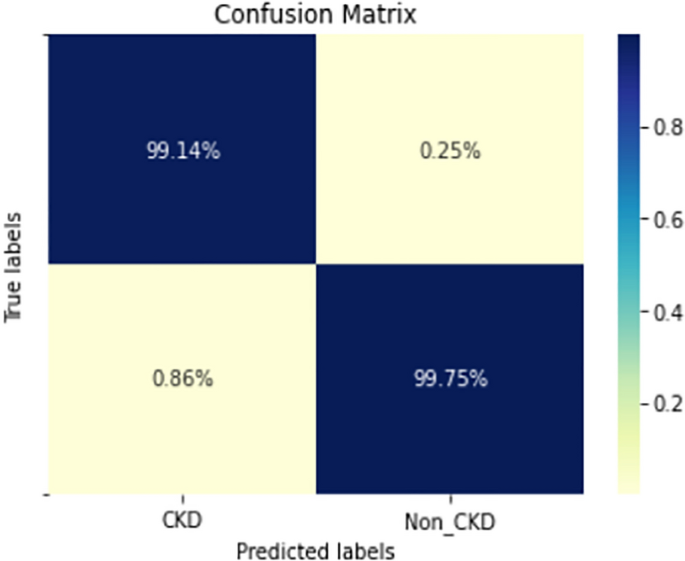
Confusion matrix of Ensemble model (12_months data)
Recently, machine learning research has shown that combining the output of several individual classifiers can reduce generalization errors and yield better performance in many applications than individual deep learning classifiers. As a result, the Ensemble learning algorithm has become dependable and dominant in a variety of fields. The Ensemble model’s main idea is to train multiple models and then combine their predictions using one of the combination techniques. In the case of kidney disease, scientists have attempted to detect it early or predict its occurrence. The practical implications of this research are that most previous studies have focused on disease detection, while only a few have focused on disease prediction before it occurs. Furthermore, the previous model’s low accuracy. Given the value of human life, developing a more accurate model leads to faster intervention to save patients’ lives.
This study focuses on Chronic Kidney Disease prediction within 6 or 12 months earlier based on medication, demographic, and comorbidity data from patients using the Deep Ensemble algorithm, which is considered a breakthrough in the scientific field. This research was conducted using two different public benchmark datasets obtained from Taiwan’s National Health Insurance Research Database. We propose three predictive models in this study: CNN model, LSTM model and the Deep Ensemble model fuses three base deep learning classifiers (CNN, LSTM, and LSTM-BLSTM) using the majority voting technique to improve classification performance.
We separately assessed the performance of the three proposed predictive models. Furthermore, we compared the proposed Deep Ensemble model’s performance to that of each base deep learning classifier. Finally, we compared the proposed models’ results to the results of the comparative paper, which used the same datasets and traditional machine learning techniques. According to the findings of this study, the proposed models performed significantly better. It can also be deduced that the proposed deep ensemble model that combines (CNN, LSTM, and LSTM_BLSTM) significantly outperforms the approaches mentioned in the literature and the deep learning models proposed in this study, with accuracy scores of 99.31%, 99.2% for 6 months and 12 months, respectively. Furthermore, the superiority of this model is distinctive.
The limitation is that this approach is appropriate for a population study but not for assisting clinicians with individual patients. To achieve the best results in patient diagnosis, decisions should be based on laboratory tests [ 13 ]. This approach, on the other hand, sheds light on previously unknown features for CKD prediction. Furthermore, due to a lack of these features in the dataset, the risk factors for this disease, such as a family history of kidney failure, hypertension, and diabetes, were not determined in this study. Finally, our deep learning models require more memory storage and a longer learning time than traditional machine learning techniques. As a result, the ensemble model needs more memory and extended time than deep learning models, as each run separately and then gathers into the proposed model.
In the future, we plan to test the robustness of our developed models against various datasets based on patient laboratory data collected from local hospitals, medical analysis laboratories, and polyclinics. Furthermore, we intend to broaden our research to include more classes of CKD detection, not just prediction, such as first and second stage CKD. Using another dataset, we can also determine the risk factors for CKD, such as a family history of kidney failure, hypertension, and diabetes.
Data availability
The datasets analysed during the current study are available in a public repository [ 23 ].
Barik S, et al. Analysis of prediction accuracy of diabetes using classifier and hybrid machine learning techniques. In: Intelligent and cloud computing. Springer; 2021. p. 399–409.
Bikku T. Multi-layered deep learning perceptron approach for health risk prediction. J Big Data. 2020;7(1):1–14.
Article Google Scholar
Alam MZ, Rahman MS, Rahman MS. A Random Forest based predictor for medical data classification using feature ranking. Inform Med Unlocked. 2019;15: 100180.
Shankar V, et al. Heart disease prediction using CNN algorithm. SN Comput Sci. 2020;1(3):1–8.
Mienye ID, Sun Y, Wang Z. An improved ensemble learning approach for the prediction of heart disease risk. Inform Med Unlocked. 2020;20: 100402.
Almansour NA, et al. Neural network and support vector machine for the prediction of chronic kidney disease: a comparative study. Comput Biol Med. 2019;109:101–11.
Kriplani H, Patel B, Roy S. Prediction of chronic kidney diseases using deep artificial neural network technique. In: Computer aided intervention and diagnostics in clinical and medical images. Springer; 2019. p. 179–87.
Jongbo OA, et al. Development of an ensemble approach to chronic kidney disease diagnosis. Sci Afr. 2020;8: e00456.
Google Scholar
Ekanayake IU, Herath D. Chronic kidney disease prediction using machine learning methods. In: 2020 Moratuwa engineering research conference (MERCon). IEEE; 2020.
Gudeti B, et al. A novel approach to predict chronic kidney disease using machine learning algorithms. In: 2020 4th international conference on electronics, communication and aerospace technology (ICECA). IEEE; 2020.
Senan EM, et al. Diagnosis of chronic kidney disease using effective classification algorithms and recursive feature elimination techniques. J Healthc Eng. 2021;2021:1–10.
Elhoseny M, Shankar K, Uthayakumar J. Intelligent diagnostic prediction and classification system for chronic kidney disease. Sci Rep. 2019;9(1):1–14.
Krishnamurthy S, et al. Machine learning prediction models for chronic kidney disease using national health insurance claim data in Taiwan. In: Healthcare. Multidisciplinary Digital Publishing Institute; 2021.
Sagi O, Rokach L. Ensemble learning: a survey. Wiley Interdiscip Rev Data Min Knowl Discov. 2018;8(4): e1249.
Zhang P, et al. Learning spatial–spectral–temporal EEG features with recurrent 3D convolutional neural networks for cross-task mental workload assessment. IEEE Trans Neural Syst Rehabil Eng. 2018;27(1):31–42.
Albawi S, Mohammed TA, Al-Zawi S. Understanding of a convolutional neural network. In: 2017 international conference on engineering and technology (ICET). IEEE; 2017.
Van Houdt G, Mosquera C, Nápoles G. A review on the long short-term memory model. Artif Intell Rev. 2020;53(8):5929–55.
Ganaie M, Hu M. Ensemble deep learning: a review. arXiv Preprint 2021. https://arxiv.org/abs/2104.02395 .
Li X, Li J. Health risk prediction using big medical data-a collaborative filtering-enhanced deep learning approach. In: 2018 IEEE 20th international conference on e-health networking, applications and services (Healthcom). IEEE; 2018.
CKD dataset-1. Available from: https://archive.ics.uci.edu/ml/datasets/chronic_kidney_disease .
Chittora P, et al. Prediction of chronic kidney disease—a machine learning perspective. IEEE Access. 2021;9:17312–34.
Singh V, Asari VK, Rajasekaran R. A deep neural network for early detection and prediction of chronic kidney disease. Diagnostics. 2022;12(1):116.
CKD-dataset-2. Available from: https://osf.io/wbv4p/?show=revision .
Atallah R, Al-Mousa A. Heart disease detection using machine learning majority voting ensemble method. In: 2019 2nd international conference on new trends in computing sciences (ICTCS). IEEE; 2019.
Neloy M, et al. A weighted average ensemble technique to predict heart disease. In: Proceedings of the third international conference on trends in computational and cognitive engineering. Springer; 2022.
Ju C, Bibaut A, van der Laan M. The relative performance of ensemble methods with deep convolutional neural networks for image classification. J Appl Stat. 2018;45(15):2800–18.
Article MathSciNet MATH Google Scholar
Mohammed A, Kora R. An effective ensemble deep learning framework for text classification. J King Saud Univ Comput Inf Sci. 2021. https://doi.org/10.1016/j.jksuci.2021.11.001 .
Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv Preprint 2014. https://arxiv.org/abs/1409.1556 .
Mahendran N, et al. Sensor-assisted weighted average ensemble model for detecting major depressive disorder. Sensors. 2019;19(22):4822.
Raza K. Improving the prediction accuracy of heart disease with ensemble learning and majority voting rule. In: U-healthcare monitoring systems. Elsevier; 2019. p. 179–96.
Yadav SS, Kadam VJ, Jadhav SM. Comparative analysis of ensemble classifier and single base classifier in medical disease diagnosis. In: International conference on communication and intelligent systems. Springer; 2019.
Asuncion A. UCI machine learning repository. University of California, Irvine, School of Information and Computer Sciences; 2007. Available from: http://www.ics.uci.edu/mlearn/MLRepository.html .
Zhou T, et al. The ensemble deep learning model for novel COVID-19 on CT images. Appl Soft Comput. 2021;98: 106885.
Chandra TB, et al. Coronavirus disease (COVID-19) detection in chest X-ray images using majority voting based classifier ensemble. Expert Syst Appl. 2021;165: 113909.
Heart disease statlog, Available from: https://archive.ics.uci.edu/ml/datasets/statlog+(heart) .
Monteral, J. C. “COVID-Chestxray database.” (2020). https://github.com/ieee8023/covid-chestxray-dataset
Alizadehsani R, et al. A database for using machine learning and data mining techniques for coronary artery disease diagnosis. Sci Data. 2019;6(1):1–13.
Hireš M, et al. Convolutional neural network ensemble for Parkinson’s disease detection from voice recordings. Comput Biol Med. 2022;141: 105021.
Lin L-Y, et al. Data resource profile: the national health insurance research database (NHIRD). Epidemiol Health. 2018;40: e2018062.
Alzubaidi L, et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. Journal of big Data. 2021;8(1):1–74.
Kaushik P, et al. EEG-based age and gender prediction using deep BLSTM-LSTM network model. IEEE Sens J. 2018;19(7):2634–41.
Bhaskar N, Suchetha M, Philip NY. Time series classification-based correlational neural network with bidirectional LSTM for automated detection of kidney disease. IEEE Sens J. 2020;21(4):4811–8.
Rahman M, Watanobe Y, Nakamura K. A bidirectional LSTM language model for code evaluation and repair. Symmetry. 2021;13(2):247.
Download references
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and affiliations.
Department of Computers and Control Engineering, Faculty of Engineering, Tanta University, Tanta, Egypt
Dina Saif, Amany M. Sarhan & Nada M. Elshennawy
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Dina Saif .
Ethics declarations
Competing interests.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Saif, D., Sarhan, A.M. & Elshennawy, N.M. Deep-kidney: an effective deep learning framework for chronic kidney disease prediction. Health Inf Sci Syst 12 , 3 (2024). https://doi.org/10.1007/s13755-023-00261-8
Download citation
Received : 27 October 2022
Accepted : 07 November 2023
Published : 01 December 2023
DOI : https://doi.org/10.1007/s13755-023-00261-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Chronic kidney disease
- CKD prediction
- Health risk prediction
- Deep learning
- Deep ensemble
- Find a journal
- Publish with us
- Track your research
Advertisement

Abstract LB391: Deep learning AI predicts HRD and platinum response from histologic slides
- Split-Screen
- Article contents
- Figures & tables
- Supplementary Data
- Peer Review
- Get Permissions
- Cite Icon Cite
- Search Site
- Version of Record April 5 2024
Erik N. Bergstrom , Ammal Abbasi , Marcos Diaz-Gay , Loïck Galland , Sylvain Ladoire , Scott M. Lippman , Ludmil B. Alexandrov; Abstract LB391: Deep learning AI predicts HRD and platinum response from histologic slides. Cancer Res 1 April 2024; 84 (7_Supplement): LB391. https://doi.org/10.1158/1538-7445.AM2024-LB391
Download citation file:
- Ris (Zotero)
- Reference Manager
Background: Cancers with homologous recombination deficiency (HRD) can benefit from platinum salts and PARP inhibitors. Standard diagnostic tests, including FDA-approved companion diagnostics, for detecting HRD require molecular profiling, which is not universally available with global testing rates lowest among minority, rural, and other underserved populations.
Methods: We trained DeepHRD, a deep-learning platform for predicting HRD from hematoxylin and eosin (H&E)-stained histopathological slides, using primary breast (n=1,008) and ovarian (n=459) cancers from The Cancer Genome Atlas (TCGA). DeepHRD was compared to four standard HRD molecular tests using breast (n=349) and ovarian (n=141) cancers from multiple external and independent datasets, including clinical cohorts with platinum complete response, progression-free survival (PFS) and overall survival (OS) endpoints.
Results: DeepHRD detected HRD from held-out H&E-stained breast cancer slides in TCGA with an AUC of 0.81 ([0.77-0.85]; 95% confidence interval). This performance was confirmed in two independent primary breast cancer cohorts (AUC=0.76; [0.71-0.82]). In an external platinum-treated metastatic breast cancer cohort, samples detected as HRD had a higher complete response (AUC=0.76; [0.54-0.93]) with 3.7-fold increase in median PFS (14.4 versus 3.9 months; p-value=0.0019) and hazard ratio (HR) of 0.45 (p=0.0047) after correcting for PAM50 molecular subtype and age at diagnosis. This deep-learning classifier outperformed four genomic HRD tests used in the clinic, including standard HRD score, BRCA1/2 , 26-HR gene panel and single-base substitution signature 3 (SBS3). Multiresolution spatial mapping identified morphological features utilized by DeepHRD for detecting HRD, notably enriched for neoplastic and necrotic tissues, and a higher macrophage density. Through transfer learning to high-grade serous-ovarian cancer, DeepHRD-positive samples exhibited better overall survival after TCGA first-line (HR=0.46; p=0.030) and an external neoadjuvant (HR=0.49; p=0.015) platinum-treated cohorts.
Conclusion: In summary, DeepHRD exhibits consistent hazard ratios ranging from 0.45 to 0.49 across the three clinical cohorts and captures 1.8- to 3.1-fold more HRD-positive breast and ovarian cancer patients. DeepHRD-positive breast cancer patients that received platinum exhibited better complete response and PFS. Similarly, DeepHRD-positive platinum-treated ovarian cancer patients had a better OS. DeepHRD’s ability to detect HRD from digital H&E slides provides an important precision oncology tool that can be utilized in resource-constrained and underserved areas where genomic testing is generally not existent.
Citation Format: Erik N. Bergstrom, Ammal Abbasi, Marcos Diaz-Gay, Loïck Galland, Sylvain Ladoire, Scott M. Lippman, Ludmil B. Alexandrov. Deep learning AI predicts HRD and platinum response from histologic slides [abstract]. In: Proceedings of the American Association for Cancer Research Annual Meeting 2024; Part 2 (Late-Breaking, Clinical Trial, and Invited Abstracts); 2024 Apr 5-10; San Diego, CA. Philadelphia (PA): AACR; Cancer Res 2024;84(7_Suppl):Abstract nr LB391.
Citing articles via
Email alerts.
- Online First
- Collections
- Online ISSN 1538-7445
- Print ISSN 0008-5472
AACR Journals
- Blood Cancer Discovery
- Cancer Discovery
- Cancer Epidemiology, Biomarkers & Prevention
- Cancer Immunology Research
- Cancer Prevention Research
- Cancer Research
- Cancer Research Communications
- Clinical Cancer Research
- Molecular Cancer Research
- Molecular Cancer Therapeutics
- Info for Advertisers
- Information for Institutions/Librarians
- Privacy Policy
- Copyright © 2023 by the American Association for Cancer Research.
This Feature Is Available To Subscribers Only
Sign In or Create an Account
- MyU : For Students, Faculty, and Staff
MCFAM Seminar - Can deep learning help solve asset-pricing equations?
Speaker: Deqing Jiang
Abstract: The curse of dimensionality stops classical methods from solving high-dimensional (differential) equations numerically. However, recent advances in deep-learning-based methods imply that these equations can be solved by reformulating them as optimization problems. In this session, we briefly review some of these deep-learning-based methods and their convergence analysis and open a discussion about how to apply them to solve (high-dimensional) asset-pricing equations.
Bio: Deqing Jiang, Final year PhD, Mathematical Institute, University of Oxford. Research direction: Analysis of deep-learning-based PDE solvers. Research interests: Mathematical finance, scientific machine learning.
- Future undergraduate students
- Future transfer students
- Future graduate students
- Future international students
- Diversity and Inclusion Opportunities
- Learn abroad
- Living Learning Communities
- Mentor programs
- Programs for women
- Student groups
- Visit, Apply & Next Steps
- Information for current students
- Departments and majors overview
- Departments
- Undergraduate majors
- Graduate programs
- Integrated Degree Programs
- Additional degree-granting programs
- Online learning
- Academic Advising overview
- Academic Advising FAQ
- Academic Advising Blog
- Appointments and drop-ins
- Academic support
- Commencement
- Four-year plans
- Honors advising
- Policies, procedures, and forms
- Career Services overview
- Resumes and cover letters
- Jobs and internships
- Interviews and job offers
- CSE Career Fair
- Major and career exploration
- Graduate school
- Collegiate Life overview
- Scholarships
- Diversity & Inclusivity Alliance
- Anderson Student Innovation Labs
- Information for alumni
- Get engaged with CSE
- Upcoming events
- CSE Alumni Society Board
- Alumni volunteer interest form
- Golden Medallion Society Reunion
- 50-Year Reunion
- Alumni honors and awards
- Outstanding Achievement
- Alumni Service
- Distinguished Leadership
- Honorary Doctorate Degrees
- Nobel Laureates
- Alumni resources
- Alumni career resources
- Alumni news outlets
- CSE branded clothing
- International alumni resources
- Inventing Tomorrow magazine
- Update your info
- CSE giving overview
- Why give to CSE?
- College priorities
- Give online now
- External relations
- Giving priorities
- Donor stories
- Impact of giving
- Ways to give to CSE
- Matching gifts
- CSE directories
- Invest in your company and the future
- Recruit our students
- Connect with researchers
- K-12 initiatives
- Diversity initiatives
- Research news
- Give to CSE
- CSE priorities
- Corporate relations
- Information for faculty and staff
- Administrative offices overview
- Office of the Dean
- Academic affairs
- Finance and Operations
- Communications
- Human resources
- Undergraduate programs and student services
- CSE Committees
- CSE policies overview
- Academic policies
- Faculty hiring and tenure policies
- Finance policies and information
- Graduate education policies
- Human resources policies
- Research policies
- Research overview
- Research centers and facilities
- Research proposal submission process
- Research safety
- Award-winning CSE faculty
- National academies
- University awards
- Honorary professorships
- Collegiate awards
- Other CSE honors and awards
- Staff awards
- Performance Management Process
- Work. With Flexibility in CSE
- K-12 outreach overview
- Summer camps
- Outreach events
- Enrichment programs
- Field trips and tours
- CSE K-12 Virtual Classroom Resources
- Educator development
- Sponsor an event
‘ICML 2023 Topological Deep Learning Challenge: Design and Results’
“This paper presents the computational challenge on topological deep learning that was hosted within the ICML 2023 Workshop on Topology and Geometry in Machine Learning. The competition asked participants to provide open-source implementations of topological neural networks from the literature by contributing to the python packages TopoNetX (data processing) and TopoModelX (deep learning). The challenge attracted twenty-eight qualifying submissions in its two month duration. This paper describes the design of the challenge and summarizes its main findings.”
Find the paper and full list of authors at Proceedings of Machine Learning Research.
‘Hierarchical RL-Guided Large-Scale Navigation of a Snake Robot’
‘bergeron: combating adversarial attacks through a conscience-based alignment framework’, ‘more samples or more prompt inputs exploring effective in-context sampling for llm few-shot prompt engineering’, ‘multi-instance randomness extraction and security against bounded-storage mass surveillance’, ‘is a seat at the table enough engaging teachers and students in dataset specification for ml in education’, ‘”the wallpaper is ugly”: indoor localization using vision and language’, ‘human still wins over llm: an empirical study of active learning on domain-specific annotation tasks’, ‘beyond labels: empowering human annotators with natural language explanations through a novel active-learning architecture’, ‘automatic collation for diversifying corpora: commonly copied texts as distant supervision for handwritten text recognition’.

IMAGES
VIDEO
COMMENTS
Deep Learning Research Proposal. The word deep learning is the study and analysis of deep features that are hidden in the data using some intelligent deep learning models. Recently, it turns out to be the most important research paradigm for advanced automated systems for decision-making. Deep learning is derived from machine learning ...
First, make sure that your proposal is well written and free of grammatical errors. Second, make sure that your proposal is clear and concise. Third, make sure that your proposal is organized and includes all of the necessary information. Finally, be sure to proofread your proposal carefully before submitting it.
The current development in deep learning is witnessing an exponential transition into automation applications. This automation transition can provide a promising framework for higher performance and lower complexity. This ongoing transition undergoes several rapid changes, resulting in the processing of the data by several studies, while it may lead to time-consuming and costly models. Thus ...
The final goal of the Artificial Intelligence community is to achieve Artificial General Intelligence, a human level of intelligence with precision, persistence, and processing speed of computers. As humans, we have the ability to progress via continual learning. We always add a skill on top of our past learned skills. Deep transfer learning, followed by progressive learning, is attempting to ...
Deep learning (DL), a branch of machine learning (ML) and artificial intelligence (AI) is nowadays considered as a core technology of today's Fourth Industrial Revolution (4IR or Industry 4.0). Due to its learning capabilities from data, DL technology originated from artificial neural network (ANN), has become a hot topic in the context of computing, and is widely applied in various ...
Thesis Proposal Scaling Distributed Machine Learning with System and Algorithm Co-design Mu Li October 2016 ... large scale machine learning has led to a surge of research interest in both academia and industry. 2. October 13, 2016 ... Take deep learning as an example, the model size in terms of the depth of the neural network has ...
The publication was founded to communicate research in a more transparent and visual way, with interactive widgets, code snippets, and animations embedded into the paper. Awesome Deep Learning Papers is a bit outdated (the last update was made two years ago) but it does list the most cited papers from 2012-2016, sorted by discipline, such as ...
Applicants hold (or are about to complete) a PhD in Operations Research, Industrial Engineering, Applied Mathematics, Electrical Engineering or a related discipline. A strong background in stochastic modeling and optimization methods is required. Research experience in statistics/machine learning/deep learning would be a great advantage.
Request PDF | On Dec 1, 2021, Mohammadreza Iman and others published EXPANSE, A Continual Deep Learning System; Research Proposal | Find, read and cite all the research you need on ResearchGate
Reading Papers: Skim: Begin with reading abstracts, introductions, conclusions, and figures. Deep Dive: When a study shows high similar to our work, then look in-depth to its methodology, experiments, and results. Take Notes: Look down the basic plans, methods, datasets, Evaluation metrics, and open issues described in the paper and note it.
This study investigated the landscape of deep learning research in biomedicine and confirmed its interdisciplinary nature. Although it has been successful, we believe that there is a need for diverse applications in certain areas to further boost the contributions of deep learning in addressing biomedical research problems. ... Proposal network ...
In the proposed research we plane to apply a deep state-of-the-art deep learning based model for BioEE. In this work, different deep learning based model structures will be designed and tested ...
This deep learning research paper looks at how TrustyAI can support trust in decision services and predictive models. The paper investigates techniques such as LIME, SHAP and counterfactuals, benchmarking both LIME and counterfactual techniques against existing implementations. ... This leads to the proposal of ABEL: an automatic scheduler ...
The application of recent artificial intelligence (AI) and deep learning (DL) approaches integrated to radiological images finds useful to accurately detect the disease. This article introduces a new synergic deep learning (SDL)-based smart health diagnosis of COVID-19 using Chest X-Ray Images. The SDL makes use of dual deep convolutional ...
A Novel Proposal for Deep Learning-Based Diabetes Prediction: Converting Clinical Data to Image Data. Muhammet Fatih Aslan, ... N.R.J. Artificial intelligence in disease diagnostics: A critical review and classification on the current state of research guiding future direction. Health Technol. 2021; 11:693-731. doi: 10.1007/s12553-021-00555-5.
To accelerate these efforts, the deep learning research field as a whole must address several challenges relating to the characteristics of health care data (i.e. sparse, noisy, heterogeneous, time-dependent) as need for improved methods and tools that enable deep learning to interface with health care information workflows and clinical ...
3.3 Deep belief networks. The Deep Belief Networks (DBN) proposed by Hinton et al. [] is a non-convolution model that can extract features and learn a deep hierarchical representation of training data.DBNs are generative models constructed by stacking multiple RBMs. DBN is a hybrid model, the first two layers are like RBM, and the rest of the layers form a directed generative model.
Abstract. I am looking for an ambitious PhD candidate in the area of Deep Learning Systems Security and Robustness.The objectives of this project are to develop new robust deep learning models ...
Although there exist a number of reviews on deep learning methods on medical image analysis [4-13], most of them emphasize either on general deep learning techniques or on specific clinical applications.The most comprehensive review paper is the work of Litjens et al. published in 2017 [].Deep learning is such a quickly evolving research field; numerous state-of-the-art works have been ...
Example research proposal #1:"A Conceptual Framework for Scheduling Constraint Management". Example research proposal #2:"Medical Students as Mediators of Change in Tobacco Use". Title page. Like your dissertation or thesis, the proposal will usually have a title pagethat includes: The proposed title of your project.
Organizing a Ph.D. proposal on deep learning. Begin by familiarizing with deep learning algorithms before writing your organize your ideas in these sections. Literature review. A literature review should provide a survey of relating works to clarify the sphere of your work. Find contemporary publications that relate to your research.
ConspectusMolecular docking, also termed ligand docking (LD), is a pivotal element of structure-based virtual screening (SBVS) used to predict the binding conformations and affinities of protein-ligand complexes. Traditional LD methodologies rely on a search and scoring framework, utilizing heuristic algorithms to explore binding conformations and scoring functions to evaluate binding ...
View PDF Abstract: In recent years there has been increased interest in understanding the interplay between deep generative models (DGMs) and the manifold hypothesis. Research in this area focuses on understanding the reasons why commonly-used DGMs succeed or fail at learning distributions supported on unknown low-dimensional manifolds, as well as developing new models explicitly designed to ...
Image created by the author using Midjourney. Parallel training on a large number of GPUs is state of the art in deep learning. The open source image generation algorithm Stable Diffusion was trained on a cluster of 256 GPUs. Meta's AI Research SuperCluster contains more than 24,000 NVIDIA H100 GPUs that are used to train models such as Llama 3.. By using multiple GPUs, machine learning ...
Abstract. Despite attempts to improve the university's training offer, dropout rates are constantly increasing. This paper is an attempt to optimize university's training offer that would ...
Abstract. Investigation of important molecular features of glioma patients may reveal driving mechanisms of patient recurrence and treatment failures in glioma. Applying various deep learning techniques enables the integration and systematic comparison of multi-omics features, allowing robust marker prediction and patient stratification. We extracted mutation and copy number variation data ...
Many researchers proposed algorithms for health risk prediction for a variety of diseases in an effort to reduce mortality. Li et al. [] forecasted the risk of hospital readmission for diabetic patients using a combination of collaborative filtering-enhanced and deep learning approaches in 2018.With 88.94% accuracy, the algorithm outperforms the Naïve Bayes, SVM, and decision tree algorithms.
This deep-learning classifier outperformed four genomic HRD tests used in the clinic, including standard HRD score, BRCA1/2, 26-HR gene panel and single-base substitution signature 3 (SBS3). Multiresolution spatial mapping identified morphological features utilized by DeepHRD for detecting HRD, notably enriched for neoplastic and necrotic ...
In this session, we briefly review some of these deep-learning-based methods and their convergence analysis and open a discussion about how to apply them to solve (high-dimensional) asset-pricing equations.Bio: Deqing Jiang, Final year PhD, Mathematical Institute, University of Oxford.Research direction: Analysis of deep-learning-based PDE ...
"This paper presents the computational challenge on topological deep learning that was hosted within the ICML 2023 Workshop on Topology and Geometry in Machine Learning. The competition asked participants to provide open-source implementations of topological neural networks from the literature by contributing to the python packages TopoNetX (data processing) and TopoModelX (deep learning). The ...